Figure 5.
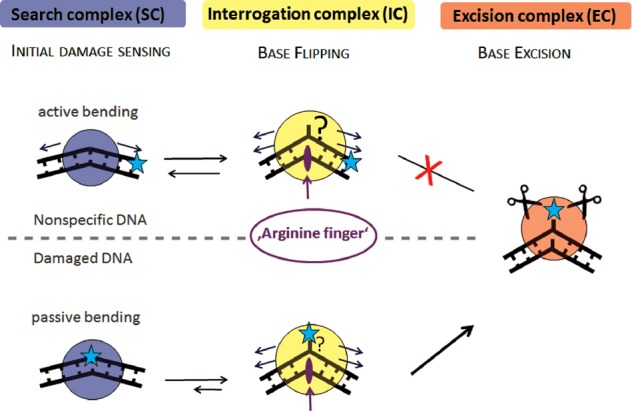
hTDG damage search model. Our proposed model for DNA lesion search and recognition by glycosylases involves three different types of TDG–DNA complexes: search complex (SC), interrogation complex (IC) and excision complex (EC). In the model, the glycosylase scans the DNA for intrinsically flexible sites, switching between an SC and an IC state. While homoduplex DNA is actively bent in the conformation of the SC (arrows indicate applied force), TDG target sites G:T and G:U form wobble base pairs (blue star) that display enhanced flexibility or intrinsic pre-bending that matches the conformation in the SC complex (passive bending). The energetic cost for DNA bending may hence serve as an initial damage-sensing mechanism that may result in a longer residence time of the glycosylase at a potential target site. In the IC, the enzyme probes base pairs via exertion of additional force on the DNA (e.g. phosphate pinching, arrows), resulting in a more strongly bent DNA structure. Protein–DNA interactions in the IC conformation may expedite spontaneous base flipping (question marks in figure) and stabilize the extrahelical base (for TDG e.g. Arg275). Correct target bases (or mismatched pairs) are then identified in the catalytic pocket of the glycosylase in the EC conformation and catalysis of base excision can occur.
