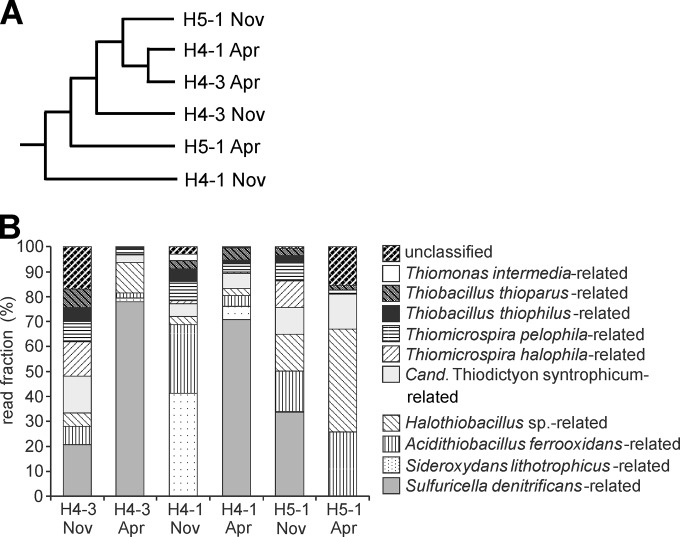
FIG 2.
Results of cbbM-targeted pyrosequencing, based on deduced cbbM protein sequences. (A) Dendrogram based on Bray-Curtis similarity values; (B) phylogenetic affiliation and relative frequency of the different sequence types. Only phylotypes represented by >1% of the sequence reads in at least one sample are shown. Closest relatives were determined based on BLAST results and distances calculated in ARB. Similarities of deduced cbbM protein sequences obtained in this study to cultivated strains are as follows: 90.4 to 97.3% for Sulfuricella denitrificans, 91.1 to 96.2% for Sideroxydans lithotrophicus ES-1, 90.5 to 97.3% for Acidithiobacillus ferrooxidans, 90.4 to 94.7% for Halothiobacillus sp., 91 to 98.7% for “Candidatus Thiodictyon,” 90.4 to 97.3% for Thiomicrospira halophila, 90.4 to 98.6% for Thiomicrospira pelophila, 90.5 to 94.6% for Thiobacillus thiophilus, 90.7 to 97.3% for Thiobacillus thioparus, and 90.4 to 97.3% for Thiomonas intermedia.