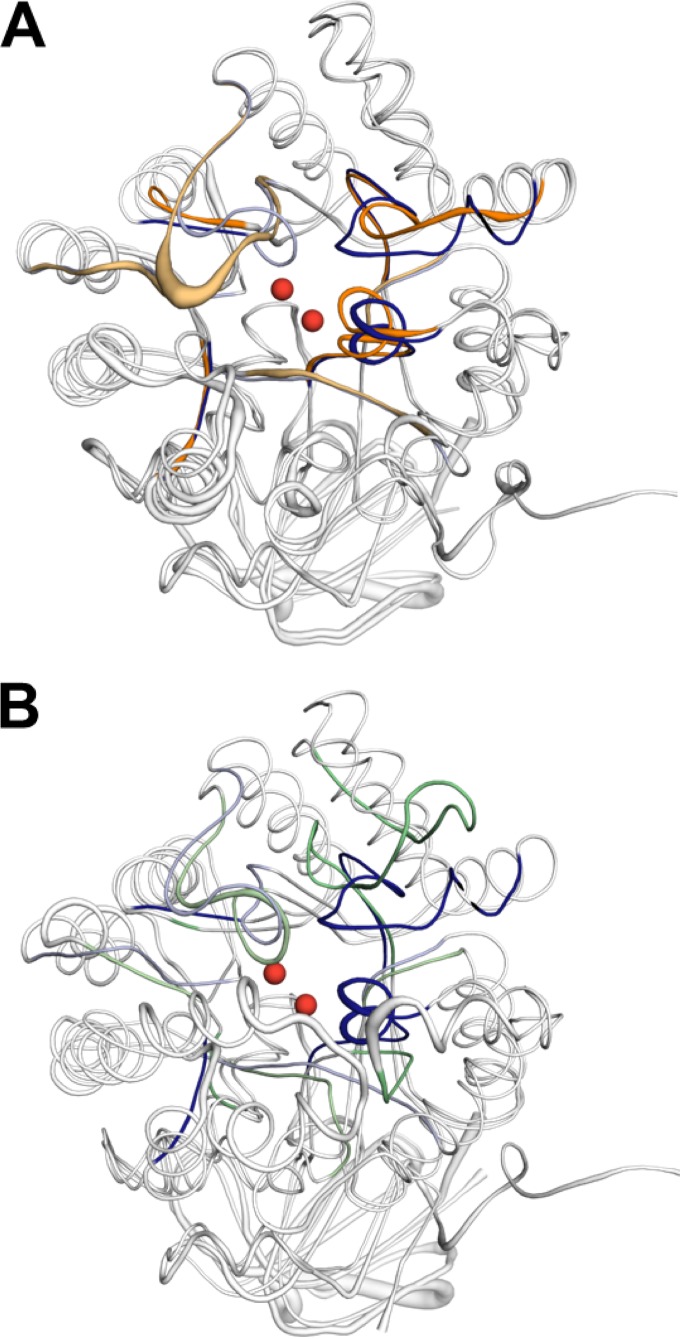
FIG 3.
(A) Loop geometry is the greatest structural difference between aligned MolA (blue) and PuhB (orange). Loops 3 and 4 exhibit the greatest divergence in geometry. The basis of the divergence in loop 3 is a three-amino-acid deletion in PuhB compared to the equivalent loop in MolA. Loop 4 has a five-amino-acid insertion in PuhB compared to MolA, which creates a significant topology difference. (B) Loop geometry is significantly different between MolA (blue) and subtype I carboxypeptidase Cc2672 (PDB code 3MTW) (green). Loops 1, 3, 4, and 8 all have very different arrangements. The two catalytic zinc ions of Cc2672 are shown as red spheres in both structures.