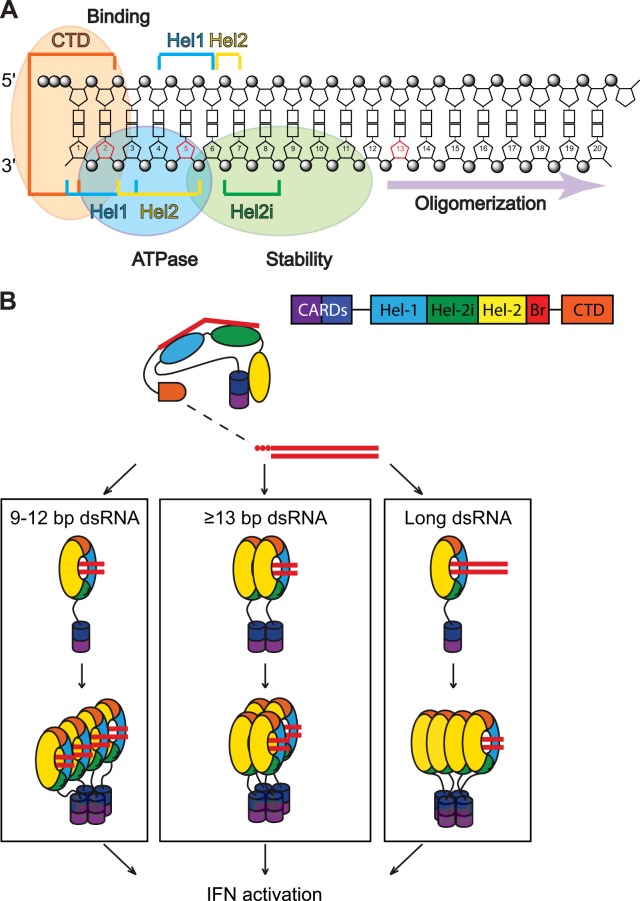
FIG 7 .
RIG-I binding and activation. (A) Schematic representation of the contacts of various RIG-I motifs to 5′ ppp-dsRNA (adapted from reference 19). Domains on the 5′ ppp-dsRNA involved in binding to RIG-I, ATPase activity of RIG-I, and stability of the RIG-I/RNA complex are indicated. (B) Model of RIG-I activation depending on the length of the dsRNA. If a tetrameric RIG-I/RNA complex is the minimal structure required for RIG-I activation (42), 3 different scenarios (1 tetramer, 2 dimers, or 4 monomers) that are dependent on the length of the dsRNA can be envisaged. The probability of forming such tetrameric complexes depends on the stability of the RIG-I/RNA complex, including the rate of recycling of RIG-I in this complex. A direct consequence of this is that it also depends on the concentration of RIG-I within the cytoplasm.