Background: Herpesviruses egress from the nucleus by vesicle trafficking through the nuclear envelope.
Results: Using giant unilamellar vesicles, we reconstitute the function of two viral proteins, pUL31 and pUL34, in nuclear envelope vesicle formation.
Conclusion: pUL34 recruits pUL31 to the membrane, which, on its own, deforms membranes to produce nuclear envelope vesicles.
Significance: pUL31 constitutes a minimal machinery mediating inwardly directed membrane budding and scission.
Keywords: Herpesvirus, Membrane Reconstitution, Membrane Trafficking, Nuclear Envelope, Nuclear Translocation
Abstract
Herpesviruses assemble capsids in the nucleus and egress by unconventional vesicle-mediated trafficking through the nuclear envelope. Capsids bud at the inner nuclear membrane into the nuclear envelope lumen. The resulting intralumenal vesicles fuse with the outer nuclear membrane, delivering the capsids to the cytoplasm. Two viral proteins are required for vesicle formation, the tail-anchored pUL34 and its soluble interactor, pUL31. Whether cellular proteins are involved is unclear. Using giant unilamellar vesicles, we show that pUL31 and pUL34 are sufficient for membrane budding and scission. pUL34 function can be bypassed by membrane tethering of pUL31, demonstrating that pUL34 is required for pUL31 membrane recruitment but not for membrane remodeling. pUL31 can inwardly deform membranes by oligomerizing on their inner surface to form buds that constrict to vesicles. Therefore, a single viral protein can mediate all events necessary for membrane budding and abscission.
Introduction
Viruses exploit amazing strategies, overcoming various cellular membranes to realize their complex life cycle, including cell entry, genome replication, and assembly of virus particles as well as their egress. For many viruses that replicate in the cell nucleus, a major barrier is the nuclear envelope, a double membrane structure enclosing the chromatin. It consists of the inner nuclear membrane, which, in animal cells, is underlaid by a tight lamina meshwork that connects and stabilizes the nuclear envelope, and the outer nuclear membrane, which is continuous with the endoplasmic reticulum (for a review, see Refs. 1, 2). The nuclear envelope is perforated by nuclear pore complexes, huge macromolecular assemblies of up to 125 MDa in vertebrates that act as selective gateways between the cytoplasm and the nucleus (3). Nuclear pores are used as entry paths for most viruses that replicate in the nucleus (for a review, see Refs. 4, 5). In many cases, this involves an at least partial disassembly of the viral capsid at the cytoplasmic side or core of the nuclear pore complex because these particles are too large to pass the pore intact.
Although many viruses assemble in the cytoplasm, herpesviruses package their newly replicated genomic DNA into capsids of up to 125 nm within the nucleus. Unlike many other DNA viruses, herpesviruses do not depend on the breakdown of the nuclear envelope during mitosis for their release and can also replicate in non-dividing cells. Therefore, they face the challenge of passing the nuclear envelope a second time, now in the opposite direction. Because herpesvirus capsids are too large for passage through the nuclear pore, they are transported through the nuclear envelope by vesicle-mediated transport, also described as an “envelopment-de-envelopment” pathway (6). Capsids bud at the inner nuclear membrane and, therefore, acquire a (primary) envelope, resulting in a nascent virus located in the lumen between the inner and outer nuclear membranes (for a review, see Refs. 7, 8). Subsequently, the primary envelope fuses with the outer nuclear membrane, resulting in translocation of the capsid to the cytoplasm. Final maturation, including assembly of the tegument and secondary envelopment, follows in the cytoplasm, and mature virions are released at the plasma membrane.
Egress from the nucleus is a multifaceted process. In all herpesviruses studied, it involves the formation of a heterodimeric nuclear egress complex consisting of two viral proteins that are conserved throughout the herpesvirus family. One component is a type II transmembrane protein, tail-anchored in the nuclear envelope and termed pUL34 in herpes simplex virus 1 and pseudorabies virus. It interacts with a soluble component, pUL31, and recruits it to the nuclear envelope (9–15). In the absence of this complex, nuclear egress is blocked, and capsids accumulate in the nucleoplasm. One function of the pUL31-pUL34 complex is the recruitment of viral and cellular kinases that phosphorylate locally and disrupt the nuclear lamina (15–19), a prerequisite for the access of capsids to the inner nuclear membrane. In addition, the pUL31-pUL34 complex might also contribute to the nuclear membrane restructuring necessary for herpesvirus nuclear egress. Transient or stable transgenic expression of pUL31 and pUL34 is sufficient to drive the formation of correctly sized primary envelopes in the perinuclear space between the outer and inner nuclear membrane in the absence of capsids (10, 20). Therefore, pUL31 and pUL34 are the only two viral proteins required for budding and fission of vesicles at the inner nuclear membrane. Whether this process is additionally dependent on the recruitment and function of cellular factors is currently unknown. Recently, it has been suggested that, in vitro, pUL31 and pUL34 are sufficient to drive membrane budding and scission by forming a coat on the surface (21). However, this assay depended on an unnaturally high content (up to 40%) of negatively charged lipid headgroups on the deformed lipid bilayer that is not present on physiological membranes. Therefore, it remains questionable to what extent it reflects the natural situation.
Primary envelopment of herpesvirus capsids requires extensive restructuring of the host inner nuclear membrane to allow envelope formation and vesicle detachment into the lumen of the nuclear envelope. Similar membrane deformations have been investigated intensively because they are at the heart of vesicle-mediated intracellular trafficking in the secretory and endocytic pathways. Many such processes are directed by complex but conceptually straightforward mechanisms in which coat proteins impose a curvature on the cytosolic membrane face, thereby generating buds that ultimately constrict to vesicles. The best known coat proteins are clathrin and the COP I and COP II complexes, which assemble on the outer side of the membrane and are crucial for vesicle formation into the cytosol (22). Membrane deformation in the opposite direction, i.e. away from the cytosol, such as the invagination of the endosomal membrane during formation of multivesicular bodies or egress of HIV and other enveloped viruses at the plasma membrane, both mediated by the ESCRT machinery, are less frequent (23). pUL31-pUL34-mediated inner nuclear membrane engulfment, much like the suggested pathway for nuclear egress of large ribonucleoprotein complexes identified recently in Drosophila (24), represents one of these exceptional pathways. It involves vesicle budding and scission from the nucleoplasm into the intermembrane space of the nuclear envelope, which is connected and topologically identical to the lumen of the endoplasmic reticulum. The molecular machinery mediating inner nuclear membrane deformation and scission remains largely obscure. It is especially unclear whether it requires proteins within the lumen of the nuclear envelope that could assemble a coat on the outer surface of the nascent vesicles, similar to COP I, COP II, and clathrin coats in the cytoplasm.
We reconstituted the function of pseudorabies virus pUL31 and pUL34 in a simple membrane system by using giant unilamellar vesicles (GUVs)2 mimicking the lipid composition of the nuclear envelope. We show that the two viral proteins are sufficient for budding and fission of membrane vesicles into the lumen of GUVs, a process that is topologically identical to inwardly directed vesicle formation at the inner nuclear membrane during herpesvirus nuclear egress. Artificial membrane recruitment of pUL31 alone results in the same membrane remodelling and generates intra-GUV vesicles. Therefore, pUL31 and pUL34 are sufficient for vesicle formation without the need for additional (cellular) proteins. Moreover, we can assign distinct functions to pUL31 and pUL34 during herpesvirus nuclear egress. pUL34 recruits pUL31 to the membrane and provides membrane anchorage, whereas pUL31 mediates membrane budding and scission.
EXPERIMENTAL PROCEDURES
1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine,4-chlorobenzenesulfonate salt (DiDC18), Alexa Fluor 546 carboxylic acid succinimidyl ester, and cascade blue-labeled neutravidin were obtained from Invitrogen, naphtopyrene from Sigma, and detergents from Calbiochem. The nuclear envelope lipid mix consisted of 5 mol% cholesterol, 2.5 mol% sphingomyeline, 2.5 mol% sodium phosphatidylserine, 10 mol% sodium phosphatidylinositol, 20 mol% phosphatidylethanolamine, and 60 mol% phosphatidylcholine (all from Avanti Polar Lipids). In lipid mixtures lacking a specific component, the respective lipid was replaced by an equimolar amount of phosphatidylcholine.
Protein Expression and Purification
Constructs for expression of pseudorabies virus pUL31 and pUL34 were generated from a synthetic DNA optimized for codon usage in Escherichia coli (Geneart). pUL31 was expressed from a modified pET28a vector with a His6 tag and a TEV site, followed by an EGFP protein amino-terminal of pUL31. The soluble domain of pUL34 (amino acids 1–240), untagged pUL31, as well as EGFP were expressed from a modified pET28a vector with a His6 tag followed by a TEV site. For C-terminal His6 tagging of the soluble domain of pUL34 (amino acids 1–240), the fragment was expressed from a modified pET28a vector lacking the amino-terminal His6 sequence. GST fusions of pUL31 and pUL34 (amino acids 1–240) were expressed from a modified pET28a vector lacking a His6 tag but with an N-terminal GST tag followed by a recognition site for precision protease upstream of pUL31 and pUL34. All His6-tagged proteins were expressed in E. coli BL21de3 and purified using Ni-NTA. Where applicable, the His6 tags were cleaved off. Proteins were purified further by gel filtration (Superdex 200, GE Healthcare). GST fusions were purified using GSH-Sepharose (GE Healthcare).
Full-length pUL34 and SCL1 were expressed from a modified pET28a vector with an N-terminal membrane-integrating sequence for translation of integral membrane protein constructs (MISTIC) fragment (25) followed by a thrombin cleavage site and purified as described previously (26). Proteins were labeled using Alexa Fluor 546 carboxylic acid succinimidyl ester in 200 mm NaHCO3 (pH 8.4) or the same buffer containing 1% (w/v) cetyltrimethylammonium bromide for labeling of transmembrane proteins and purified by gel filtration on a Sephadex G50 fine column (GE Healthcare).
Generation of GUVs
Detergent-solubilized and labeled SCL1 and pUL34 were reconstituted in proteoliposomes via gel filtration (27). For this, 20 μl of the nuclear envelope lipid mix (30 mg/ml in 10% octylglucopyranoside) was mixed with 20 μl of 2 μm pUL34 or SCL1 protein and 100 μl of PBS. The sample was applied to a Sephadex G50 fine filled Econo chromatography column (0.5 × 20 cm, Bio-Rad) to remove the detergent. The formed proteoliposomes were collected and pelleted for 30 min at 100,000 rpm in a TLA120.2 (Beckman Coulter) rotor at 4 °C. The pellet was resuspended in 120 μl of 20 mm HEPES (pH 7.4), 100 mm KCl, and 1 mm DTT. 5 μl of resuspended proteoliposomes were dried onto two 5 × 5 mm platinum gauzes (ALS) under vacuum for at least 1 h at room temperature. The gauzes were placed in parallel (5 mm distance) into a cuvette (UVette, Eppendorf) and submerged in 259 mm sucrose solution, and for 140 min an alternating current electric field with 10 Hz, 2.2 V was applied, followed by 20 min at 2 Hz at 42 °C.
Lipid GUVs were generated from chloroform-dissolved lipid mixtures where indicated, supplemented with 1 mol% Ni-NTA-DGS (1,2-dioleoyl-sn-glycero-3-(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl), Avanti Polar Lipids) and 0.8 nm DiDC18 as described previously (28).
GUV Vesicle Budding Reaction
An 8-well glass observation chamber (chambered #1.0 borosilicate coverglass system, Lab-Tek) was blocked with 5% (w/v) BSA in PBS and washed with PBS. For each reaction, 50 μl of freshly prepared GUVs were mixed with 150 μl of PBS and placed into a well. Unless indicated otherwise, all soluble proteins were added to a final concentration of 500 nm (cascade blue-labeled neutravidin at 3.8 μm). The proteins and buffers used for GUV preparation matched the osmotic pressure of the sucrose solution, as measured using an osmometer. The mixture was incubated for 5 min and imaged immediately at room temperature on an inverted Olympus Fluoview 1000 confocal laser-scanning system utilizing an UPlanSApo ×60/1.35 oil objective. The cascade blue-labeled neutravidin was excited by a 405-nm diode pumped solid state laser, and emission was collected between 425–475 nm. EGFP and Alexa Fluor 546 were excited by an argon ion laser at 488 and 515 nm, respectively. Emission for EGFP was collected between 500–545 nm and between 570–625 nm for Alexa Fluor 546. DiD was excited by a 635-nm DPSS laser, and emission was collected between 655–755 nm. The pinhole was set to 1 airy unit. Three-dimensional reconstructions were generated using Imaris software (version 7.4, Bitplane). Linescan analysis was performed using FIJI software (Fiji.sc/Fiji). Size quantification was done by stack analysis of GUVs using a stack size of 0.5 μm.
Preparation of Supported Lipid Bilayers
Supported lipid bilayers were prepared as described previously (29). The nuclear envelope lipid mix with 0.2 mol % Rhodamine-PE was dissolved in SLB buffer (10 mm HEPES (pH 7.4) and 150 mm NaCl) at a concentration of 5 mg/ml. 20 μl of the mixture was diluted with 150 μl of SLB buffer. The suspension was then vortexed and bath-sonicated until a clear suspension was obtained, indicating that small unilamellar vesicles had formed. The clear solution was put in contact, at 37 °C, with freshly cleaved mica glued to a coverslip. CaCl2 was added to a final concentration of 3 mm and incubated at 37 °C for 10 min. The samples were rinsed several times with SLB buffer to remove CaCl2 and unfused vesicles and then allowed to equilibrate at room temperature before analysis.
Confocal Microscopy of Supported Lipid Bilayers
Supported lipid bilayers were imaged using a commercial LSM 710 (Carl Zeiss, Jena, Germany) at 22 °C. The excitation light of a DPSS laser at 561 nm and of an argon laser at 488 nm was reflected by a dichroic mirror (main beam splitter 488/561/633) and focused through a Zeiss C-Apochromat ×40, numerical aperture 1.2 water immersion objective onto the sample. The fluorescence emission was collected by the objective and directed by spectral beam guides to photomultiplier tube detectors. Images were acquired before and adding His6-EGFP-pUL31 or His6-EGFP at the same areas of the supported lipid bilayers. Images were analyzed using FIJI (particle analysis). Proteins were added to a final concentration of 500 nm.
Atomic Force Microscopy
Supported lipid bilayers were imaged using a JPK NanoWizard II system (JPK Instruments, Berlin, Germany) mounted on an Axiovert 200 inverted microscope (Carl Zeiss). Intermittent contact (or tapping) mode images were taken using V-shaped silicon nitride cantilevers (SNL-10) with a typical spring constant of 0.08 N/m and a nominal tip radius of 2 nm. The cantilever oscillation was tuned to a frequency between 3–10 kHz, and the amplitude was set between 0.3–0.6 V. The amplitude was varied during the experiment to minimize the force of the tip on the bilayer. The scan rate was set to 0.7–1 Hz. The height, deflection, and phase shift signals were collected simultaneously in both trace and retrace directions. Atomic force microscopy images were acquired before and after adding 150 nm His6-EGFP-pUL31 or His6-EGFP on the bilayer. Atomic force microscopy images were analyzed using JPK data analysis software.
RESULTS
pUL31 and pUL34 Are Sufficient for Membrane Budding and Scission
Viral pUL31 and pUL34 are necessary for nuclear egress of herpesvirus capsids. To investigate their function in detail, we expressed the transmembrane protein pUL34 in E. coli as a full-length protein, including its transmembrane region, and, after purification, labeled it with the fluorescent dye Alexa Fluor 546 before reconstitution into liposomes with a nuclear envelope-like lipid composition (30, 31) (see “Experimental Procedures” for details). GUVs were generated from these proteoliposomes. The GUV membrane contained pUL34 as an integral membrane component, which can be detected by its fluorescent label (Fig. 1A). The vast majority of these GUVs were large vesicles with no detectable membrane/vesicle structures in the interior.
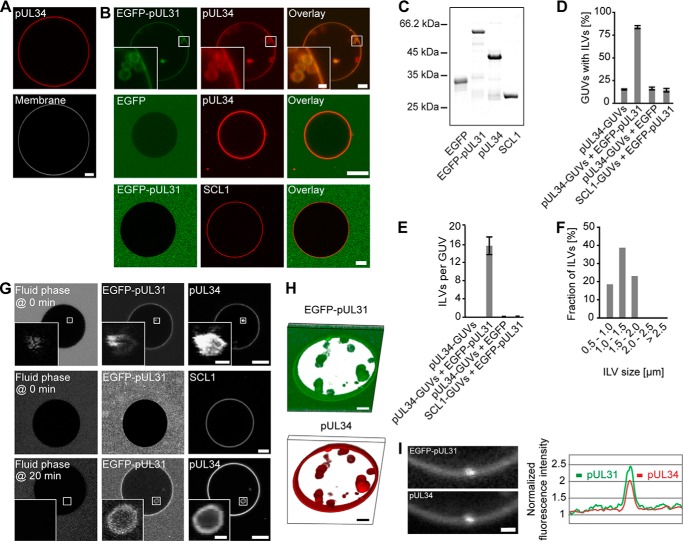
FIGURE 1.
The pUL31-pUL34 complex is sufficient to induce intralumenal vesicles. A, recombinant Alexa Fluor 546-labeled pUL34 (red) was reconstituted into GUVs. The GUV membrane was stained with the lipophilic dye DiDC18. B, Alexa Fluor 546-labeled pUL34 or the inner nuclear membrane protein SCL1 were reconstituted into GUVs (red). Addition of EGFP-pUL31 (green in overlay) induced formation of intra-GUV vesicles on pUL34 but not SCL1 GUVs. C, recombinant proteins employed were separated on 12% SDS-PAGE and stained with Coomassie Blue. D, quantitation of the number of GUVs with intralumenal vesicles (ILVs) shows the mean ± S.E. of three independent experiments, each including at least 70 GUVs/condition and experiment. E, the number of ILVs per GUV was quantified from three independent experiments, each including at least 20 GUVs/condition and experiment. The mean ± S.E. is shown. F, the size distribution of ILVs formed in pUL34-GUVs after EGFP-pUL31 addition was analyzed from three independent experiments, each including at least 20 GUVs/condition and experiment. G, cascade blue-labeled neutravidin (fluid phase marker) was added together with EGFP-pUL31 (top row) or 20 min after EGFP-pUL31 addition to GUVs (bottom row) with reconstituted Alexa Fluor 546-labeled pUL34 or SCL1. H, three-dimensional reconstruction of an EGFP-pUL31-treated (green) pUL34 GUV (red). I, higher magnification of a budding spot on a pUL34 GUV after EGFP-pUL31 addition. EGFP-UL31 and Alexa Fluor 546-pUL34 was analyzed along the limiting GUV membrane. Scale bars = 5 μm (1 μm in I and insets).
When added to pUL34-GUVs, purified recombinant EGFP-pUL31 was efficiently recruited to the membranes (Fig. 1B), consistent with a direct pUL31-pUL34 interaction (9, 12, 14). In the presence of pUL31, the GUV membrane was deformed and invaginated, leading to small intralumenal vesicles inside the GUVs that contained both pUL31 and pUL34. In typically sized GUVs with a diameter of 30–40 μm, ∼15 intralumenal vesicles were detected, the majority of them 0.5–2 μm in size (Fig. 1, D–F). Formation of intralumenal vesicles was specific for pUL31 because it was not induced by addition of EGFP alone. The same result was observed in different buffer systems and with untagged pUL31, confirming that the EGFP tag did not cause this effect.3 As a control, we replaced pUL34 with the inner nuclear membrane protein SCL1/BC08 (32). Like pUL34, SCL1 is a type II transmembrane protein with a single membrane-spanning domain at its C terminus. Therefore, the N-terminal region preceding the transmembrane domain of both pUL34 and SCL1 faces the nucleoplasm. EGFP-pUL31 was not recruited to SCL1-GUVs, and membrane invaginations were not observed above background levels, demonstrating that this process specifically requires pUL31 and pUL34 (Fig. 1B).
Cascade blue-labeled neutravidin was used as a fluid phase marker to assess permeability between the bulk solution and the pUL31-induced vesicles. When the fluid phase marker was added as a soluble probe, together with pUL31, to pUL34-GUVs, the label was found in intralumenal vesicles (Fig. 1G, top row). When added 20 min after pUL31 addition, intra-GUV vesicles without the fluid phase marker were detected (Fig. 1G, bottom row). Therefore, the lumina of vesicles are disconnected from the bulk solution surrounding the GUVs, indicating membrane scission. This is consistent with the three-dimensional reconstitution of an EGFP-pUL31-treated pUL34-GUV that shows highly mobile vesicles distant and apparently detached from the limiting GUV membrane (Fig. 1H). When the fluid phase marker was added to SCL1-GUVs together with EGFP-pUL31 (Fig. 1G) or to pUL34-GUVs in the absence of EGFP-pUL31,3 no intra-GUV fluorescence was detected, consistent with the notion that the pUL31-pUL34 interaction specifically induces vesicle budding into the GUV lumen.
These data show that, in a minimal membrane system, the two viral proteins pUL31 and pUL34 are sufficient to induce membrane perturbations generating intra-GUV vesicles. Importantly, in our assay, pUL34 is integrated as a transmembrane protein into the GUV lipid bilayer, mimicking the natural situation at the nuclear envelope membranes. The restructuring of the GUV-limiting membrane is reminiscent of the inwardly directed primary vesicle formation into the lumen of the nuclear envelope during herpesvirus nuclear egress. The emerging buds were enriched in pUL31 and pUL34, indicating that both proteins accumulate at sites of membrane deformation (Fig. 1I), consistent with a role in this process. Therefore, this GUV system is a valuable tool to investigate pUL31-pUL34-mediated vesicle budding and scission in detail.
pUL31 Mediates Membrane Budding and Scission
Many membrane-deforming processes, including budding of several viruses at the plasma membrane, specifically involve integral membrane proteins (33–37). To assess whether the membrane-spanning C terminus of pUL34 is required for the formation of intralumenal vesicles, we artificially recruited a pUL34 fragment lacking its transmembrane domain to GUVs (Fig. 2A). To this end, the Alexa Fluor 546-labeled soluble domain (amino acids 1–240) of pUL34 fused to an N-or C-terminal His6 tag was added to GUVs incorporating the Ni2+-chelating lipid Ni-NTA-DGS. GUVs coated with these tethered pUL34 fragments showed no detectable vesicle structures in their interior. When EGFP-pUL31 was added, it was efficiently recruited to the GUV membranes regardless of whether pUL34 was attached to the membrane via its N or C terminus. This finding is consistent with the fact that the transmembrane region of pUL34 is not required for pUL31 interaction (9, 38). More importantly, pUL34-mediated pUL31 recruitment was sufficient for intralumenal vesicle formation. This indicates that the presence of an authentic transmembrane region is not essential for this process (21).
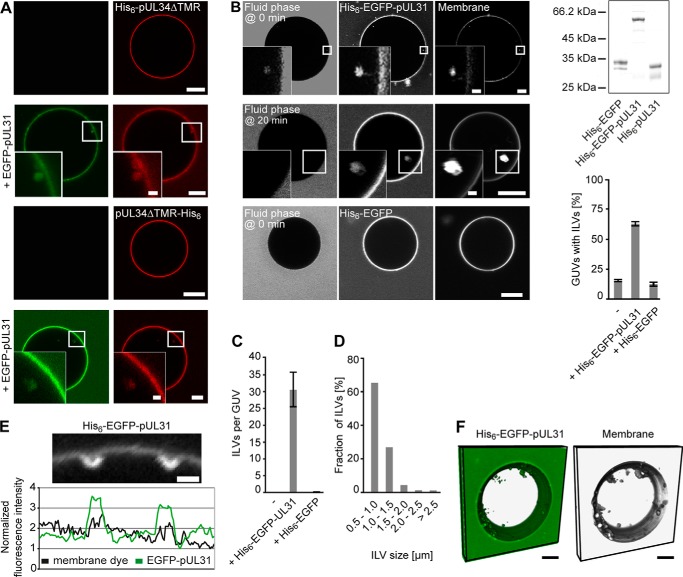
FIGURE 2.
Membrane tethering of pUL31 is sufficient to induce intralumenal vesicles. A, an Alexa Fluor 546-labeled N- or C-terminally His6-tagged pUL34 fragment (top and bottom rows, respectively) comprising the soluble domain (amino acids 1–240) was bound to 1% Ni-NTA-DGS-containing GUVs. Addition of EGFP-pUL31 induced intra-GUV vesicles. B, His6-tagged-EGFP or His6-tagged-EGFP-pUL31 was directly bound to Ni-NTA-DGS-containing GUVs. Membranes were stained with DiDC18. Cascade blue-labeled neutravidin as a fluid phase marker was added together with His6-tagged EGFP-pUL31 or His6-tagged-EGFP or 20 min after His6-EGFP-pUL31 addition to GUVs. The purity of the employed recombinant proteins is shown by SDS-PAGE and Coomassie staining. Quantitation shows the mean ± S.E. of three independent experiments, each including at least 80 GUVs/condition and experiment. C, the number of ILVs per GUV was quantified from three independent experiments, each including at least 20 GUVs/condition and experiment. The mean ± S.E. is shown. D, the size distribution of ILVs formed in Ni-NTA-DGS-containing GUVs after His6-EGFP-pUL31 addition was analyzed from three independent experiments, each including at least 20 GUVs/condition and experiment. E, higher magnification visualizing budding spots on His6-EGFP-pUL31-tethered Ni-NTA-DGS GUVs. F, three-dimensional reconstruction of a His6-EGFP-pUL31-treated (green) GUV containing 1% Ni-NTA-DGS. Membranes were stained with DiDC18 (gray). Scale bars = 5 μm (1 μm in E and insets).
It should be noted that, in contrast to the experiments presented in Fig. 1, where pUL34, because of its reconstitution as an integral membrane protein, might be present in both orientations in the GUV membrane, the pUL34 fragment is only located on the outer leaflet of the GUVs. Although unlikely in the light of its cellular localization, orienting its non-membrane-spanning region into the nucleoplasm, the experiments shown here exclude that a (wrongly oriented) fraction of pUL34 is causative for in vitro vesicle formation.
Next we tested whether pUL34 plays an active role in the invagination process or whether it is merely required for pUL31 recruitment. When His6-tagged EGFP-pUL31 was bound directly to Ni-NTA-DGS-containing lipid GUVs, intralumenal vesicles were induced efficiently induced (Fig. 2, B and C). Addition of cascade blue-labeled neutravidin together or 20 min after protein addition indicated that the formed intralumenal vesicles pinched off from the limiting GUV membrane. The size distribution of these intralumenal vesicles was shifted to smaller vesicle diameters (Fig. 2D). Although we cannot exclude other scenarios, it is possible that this is due to the absence of pUL34. Three-dimensional reconstitution of a His6-EGFP-pUL31-treated GUV shows many highly mobile vesicles distant from the GUV membrane, indicating that they have been disconnected (Fig. 2F). Therefore, artificial membrane tethering of pUL31 is sufficient for the induction of membrane invaginations and membrane scission. This effect was specific for pUL31 because a His6-tagged EGFP, despite efficient recruitment to the GUV membrane, did not induce intra-GUV vesicle formation (Fig. 2, B and C). Therefore, the function of pUL34 in this minimal system is to target pUL31 to the membrane.
pUL31-pUL34-mediated Membrane Resculpting Requires Cholesterol and Sphingomyelin
The GUVs used so far contained a lipid composition resembling the nuclear envelope. We next assessed whether any specific lipid or lipid class in this mixture is required for intralumenal vesicle formation. GUVs were reconstituted with full-length pUL34 in the absence of specific lipid components. EGFP-pUL31 was efficiently recruited to the membrane of pUL34-GUVs regardless of lipid composition (Fig. 3A). In fact, efficient formation of intralumenal vesicles was observed in all instances, except when cholesterol or sphingomyelin were absent from the GUV membrane. Also, in the absence of both negatively charged lipid classes, phosphatidylserine and phosphatidylinositol, intralumenal vesicles formed efficiently. Identical results were obtained when His6-tagged EGFP-pUL31 was directly targeted to Ni-NTA-DGS GUVs (Fig. 3B). These data indicate that cholesterol and sphingomyelin are required for pUL31-induced membrane invagination and scission. Both lipids interact and are crucial for lateral segregation of membrane components, leading to membrane subdomain formation, such as rafts (39). However, we did not observe a pUL31-pUL34-induced phase separation of sphingolipid-cholesterol from other membrane components in GUVs (Fig. 3C), although we cannot exclude the formation of raft-like nanodomains of a size below the optical resolution of light microscopy. It is possible that cholesterol and sphingomyelin are required for intra-GUV vesicle formation because they permit membrane fluidity (40) and, therefore, might be essential for flexibility of the lipid bilayer, allowing its resculpting to vesicles.
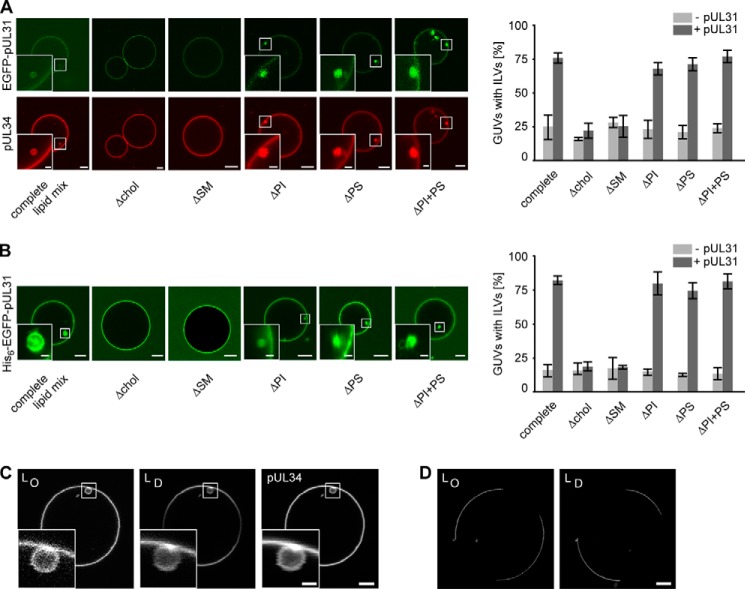
FIGURE 3.
Cholesterol and sphingomyelin are required for pUL31-mediated vesicle formation. A, recombinant Alexa Fluor 546-labeled pUL34 was reconstituted into GUVs containing the nuclear envelope lipid mix (complete lipid mix) or the same lipid mix lacking either cholesterol (chol), sphingomyelin (SM), the negatively charged phospholipids phosphoinositol (PI) or phosphatidylserine (PS), or both (PI+PS). Formation of intralumenal vesicles was induced and quantified after EGFP-pUL31 addition (mean ± S.E. of three independent experiments, each including at least 50 GUVs/experiment and condition). B, His6-tagged-EGFP-pUL31 was bound directly to Ni-NTA-DGS-containing GUVs with the same lipid compositions as in A, and ILV numbers were quantified (mean ± S.E. of three independent experiments, each including at least 60 GUVs/experiment and condition). C, pUL31 was added to pUL34-GUVs loaded with the membrane dyes naphthopyrene and DiDC18, which label the liquid ordered (LO) or liquid disordered phase (LD), respectively. Although we cannot exclude an enrichment of specific lipids in internal vesicles, there no induction and separation of liquid-ordered and -disordered phases was detectable. D, as a control for the functionality of the membrane dyes, their segregation was tested on phase-separating membrane GUVs (33 mol% cholesterol, 33 mol% sphingomyelin, 33 mol% phosphatidylcholine). Scale bars = 5 μm (1 μm in insets).
pUL31 Oligomerizes on the Membrane Surface
Our data show that pUL31 membrane recruitment induces vesicle formation on GUVs. Many membrane remodelling proteins, such as clathrin or components of the COP I/II or ESCRT machinery, oligomerize on the deforming membrane (22, 23). His6-EGFP-pUL31 became enriched at the sites of inwardly directed membrane deformation (Fig. 2E). To test for membrane-induced oligomerization, we bound limiting amounts of His6-pUL31 to Ni-NTA-DGS GUVs. Under these conditions, no intralumenal vesicles were detected (Fig. 4). Addition of increasing amounts of EGFP-pUL31 devoid of a His6 tag resulted in recruitment of the protein to the membrane and induction of intralumenal vesicle formation. EGFP-pUL31 membrane recruitment was mediated specifically by GUV tethered His6-pUL31 because EGFP-pUL31 membrane labeling and vesicle formation was not observed in the absence of His6-pUL31. Interestingly, pUL31 does not self-interact in solution when tested by GST pulldown experiments or gel filtration followed by multiangle laser light scattering (Fig. 4, B–D).
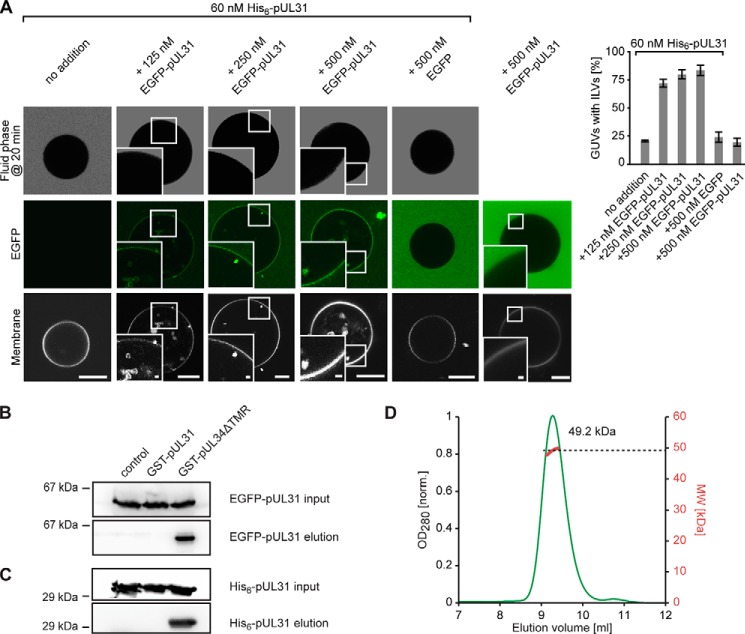
FIGURE 4.
pUL31 self-interacts on membranes but not in solution. A, where indicated, 60 nm His6-tagged-pUL31 was bound directly to Ni-NTA-DGS-containing GUVs. Increasing amounts of EGFP-pUL31 were added to the GUVs showing a pUL31-mediated membrane recruitment of EGFP-pUL31 and formation of intra-GUV vesicles. Where indicated, cascade blue-labeled neutravidin was added as a fluid phase marker 20 min after protein addition to GUVs to confirm scission of ILVs from the limiting GUV membrane. Quantitation shows the mean ± S.E. of three independent experiments, each including at least 60 GUVs/condition and experiment. Scale bars are 10 μm (1 μm in insets). B, GST pulldown using GST (control), GST-pUL31, and GST-pUL34ΔTMR (amino acids 1–240, i.e. lacking the transmembrane region) as bait and EGFP-pUL31 as prey. GST-bound EGFP-pUL31 was eluted with precision protease-containing buffer, which cleaves the GST fusions C-terminal of the GST tag, as analyzed by Western blotting using anti-EGFP-antibodies. C, GST pulldown as in B, but with His6-pUL31 as prey, detected using anti-His6 antibodies. D, size exclusion chromatography on a Superdex 75/300 GL column followed by multiangle static laser light scattering of EGFP-UL31 shows that it is monomeric in solution (calculated mass, 58.7 kDa). The red dots relate to the secondary axis and show the molecular weight of the eluting particle.
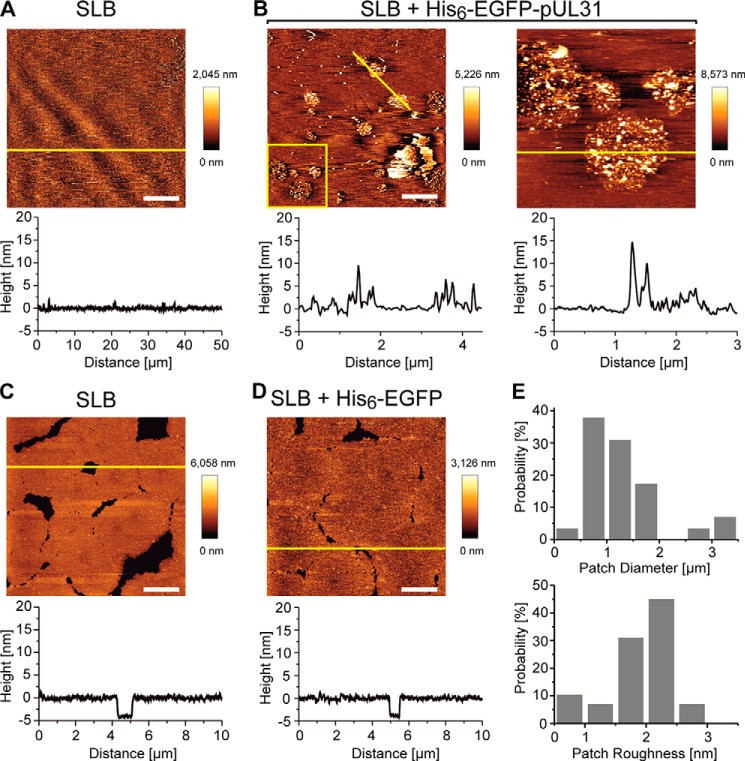
To directly observe pUL31 oligomerization on the membrane surface and induction of membrane remodelling activity, we incubated the protein with supported lipid bilayers of nuclear envelope lipid composition supplemented with Ni-NTA-DGS. His6-EGFP-pUL31 was recruited rapidly to the supported lipid bilayers, where it formed patches of 1.0 ± 0.5 μm in diameter (Figs. 5 and 6) and eventually provoked the appearance of multiple defects, indicating disruption of the lipid bilayer (Fig. 6). This latter effect is likely due to the increase in membrane tension associated with the membrane deformations at the pUL31 assembly sites. In contrast, His6-EGFP was also recruited efficiently to the supported lipid bilayers but did not induce patch formation or disruption of the bilayer.
FIGURE 5.
pUL31 forms patches on supported lipid bilayers. A, supported lipid bilayers (SLB) mimicking the nuclear envelope lipid composition (supplemented with Ni-NTA-DGS) show a flat topography before protein addition. Graphs below the image are profiles of the yellow line in the image. B, upon addition of His6-EGFP-pUL31, patches form with aggregated structures. The right figure is a 3 × 3 μm enlarged image of the yellow box within the left panel. C, supported lipid bilayers before addition of His6-EGFP. D, supported lipid bilayers after addition of His6-EGFP with no change in topography detectable. E, diameter and roughness (average height of the patches) distributions of the patches observed upon addition of His6-EGFP-pUL31. Gaussian fitting of the histograms shows a peak 1.0 ± 0.5 μm in diameter (mean ± S.D.) and 2.0 ± 0.1 nm in roughness. Scale bars = 10 μm for A and 2 μm for B–D.
FIGURE 6.
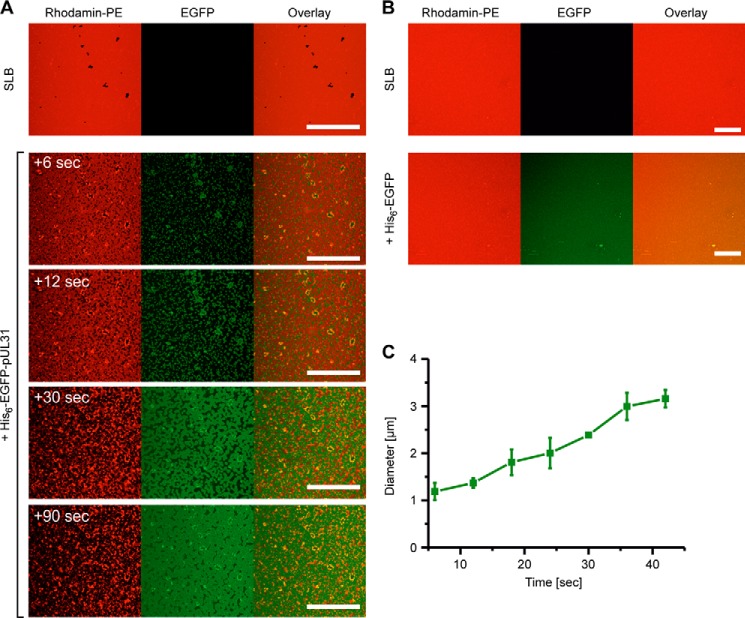
pUL31 disrupts supported lipid bilayers. A, supported lipid bilayers (SLB) mimicking the nuclear envelope with rhodamine-PE (red) form continuous fluid phase. Bilayers are disrupted upon addition of His6-EGFP-pUL31 (green). A few seconds after addition, pUL31 forms 1.2 ± 0.2 μm patches that grow over time until they cover the whole area (around 90 s). The bilayer is destroyed, and lipid aggregation is seen over this period of time. B, addition of His6-EGFP did not change the bilayer. C, particle analysis of the patches showing the growth of the particles over time. After 40 s, particles fuse and can no longer be analyzed. Scale bars = 50 μm.
Together, these data indicate that pUL31 forms clusters when recruited to membranes. Therefore, our data suggest that when monomeric pUL31 is recruited to membranes in the cellular context by pUL34, it self-interacts to induce membrane deformation, leading to vesicle formation.
DISCUSSION
Nuclear egress is a common mechanism for herpesvirus nucleocapsid translocation through the nuclear envelope (41). Although it has long been thought to be unique to herpesviruses, it has recently been recognized that a similar mechanism also functions in the nuclear export of large ribonucleoprotein particles in Drosophila (24), indicating the presence of a general, so far unknown mechanism of vesicular transport through the nuclear envelope. Although the cellular factors involved in this process remain largely enigmatic but may include the AAA+ ATPase torsin (42), all members of the Herpesviridae family analyzed in this respect are dependent on the viral nuclear egress complex, composed of pUL31 and pUL34 homologs, for translocation through the nuclear envelope. In the absence of either complex partner, herpesvirus-induced nuclear envelope breakdown can substitute for pUL31-pUL34-mediated nuclear egress (43). This mode of nuclear escape has, however, only been observed after forced reversion analysis in cell culture. Differing roles for pUL31 and pUL34 in nuclear egress have been demonstrated (7, 8), including recruitment of viral and cellular kinases that locally disrupt the lamina (15–19) as well as functions in herpesvirus replication beyond nuclear egress, including viral DNA binding and packaging (44, 45) or cell-to-cell spread (46).
Here we reconstituted herpesvirus pUL31-pUL34-dependent membrane invagination and scission using a minimal set of protein components and a simple model for the eukaryotic nuclear envelope. The presence of pUL31 at the membrane is sufficient to induce GUV-internal vesicles, a process topologically similar to the formation of primary envelopes, indicating that, in this system, pUL34 is only required to recruit pUL31 to the membrane. In contrast to a recent report (21), we did not observe a dependence on the negatively charged phospholipids phosphatidylserine and phosphatidic acid, which needed to be present in unphysiologically high amounts (up to 40%). In our system, budding and scission also occur in the absence of the comparably low amounts of phosphatidylserine and phosphatidylinositol found in the nuclear envelope. This difference is likely due to the fact that, in the previous report, the negatively charged lipids are required for membrane recruitment by electrostatic interaction of a C-terminally truncated pUL34 lacking the transmembrane region. In contrast, we used either full-length pUL34 containing the C-terminal transmembrane region or directly tethered pUL31 to the GUV membrane. Therefore, our system is less prone to artifacts and mimics the natural situation more faithfully.
The minimal GUV model system provides several fundamental new insights into the process of herpesvirus nuclear egress. First, it shows that pUL31-pUL34-mediated membrane deformation at the inner nuclear membrane can be uncoupled and is, therefore, functionally disconnected from lamina disassembly. It is formally possible that pUL31-pUL34 regulated lamina dynamics are a driving force for restructuring the inner nuclear membrane, similar to other cytoskeleton assembly and disassembly processes that are linked to membrane shape changes of a variety of organelles (47, 48). However, in our minimal system, deformation of the GUV membrane leading to vesicle formation occurs in the absence of lamin proteins. Second, pUL31 and pUL34 have well defined and separate functions in the primary envelopment of herpesvirus capsids. pUL31 and pUL34 are both crucial for this process, and coexpression of both proteins in uninfected cells induces the formation and scission of vesicles from the inner nuclear envelope (10, 20). It could be envisioned that cellular proteins participate in the necessary restructuring of the inner nuclear membrane because a similar process occurs during the nuclear export of large ribonucleoprotein particles in Drosophila neurons (24). However, our work shows that cellular proteins are not essential to execute the basic membrane restructuring necessary for nuclear egress, including membrane deformation, budding away from the nucleoplasm, and scission to generate vesicles detached from the inner nuclear membrane. In contrast, cellular proteins may be involved in fusion of the intralumenal vesicles with the outer nuclear membrane to complete the transport process. Third, pUL31 membrane recruitment is sufficient for membrane remodelling, resulting in vesicle formation. Therefore, this viral protein is the driving force both for membrane budding and scission in herpesvirus nuclear egress, which constitutes a new archetype of these non-conventional, inwardly directed budding events.
Herpesvirus nuclear egress can be regarded as vesicle-mediated transport through the lumen of the nuclear envelope. In this respect, the process has an inverted topology compared with classical vesicular trafficking pathways through the cytosol. The prime factors inducing membrane deformation and scission in vesicular trafficking localize and act on the outer surface of the exvaginated membrane and/or the vesicle. Our results demonstrate that such an outer membrane coat is not required for vesicle formation during herpesvirus nuclear egress. Rather, formation of the nuclear egress complex on the emerging inner vesicle surface is sufficient to drive membrane bending and scission. Different mechanisms implicated in membrane remodelling might be envisioned to promote vesicle formation (35). First, integral membrane proteins can deform membranes. Well known examples are reticulons and caveolins, which oligomerize and possess unusual hydrophobic segments that might form wedge-shaped hairpins in the membrane (33, 34), and both features can contribute to membrane shaping (35). Similarly, budding of some viruses (e.g. coronaviruses) at the plasma membrane is driven by membrane protein oligomerization (49). However, sequence analysis does not suggest such an unusual topology for the pUL34 transmembrane region. Our data rather show that pUL34 is dispensable for membrane restructuring leading to vesicle formation in the nuclear envelope lumen. Furthermore, membrane invaginations can be detected in the GUV system with pUL31 alone, indicating that a transmembrane region does not play a compulsory role in the process.
A second widely discussed mechanism that can contribute to membrane deformation and budding is the insertion of amphipathic helixes into the lipid bilayer (31, 50, 51). However, neither pUL31 nor pUL34 are predicted to form such helixes. More importantly, helix insertion has to take place in the outer lipid leaflet of the nascent vesicle to increase the outer in relation to the inner surface area. pUL31 is localized in the interior of the vesicle, discounting such a mechanism. Similarly, protein crowding generating lateral pressure, which can curve membranes (52), can be excluded for pUL31-mediated membrane budding because this mechanism generates membrane deformation in the opposite direction, i.e. outward from the limiting membrane.
In addition to protein-driven processes, lipid-induced changes can restructure membranes. Phase separation can generate membrane curvature and induce budding and scission in simple membrane systems (53–55). The membrane envelope of HIV is highly enriched in the raft-forming lipids sphingomyelin and cholesterol (56, 57), which suggests that budding at the plasma membrane occurs from lipid rafts (58). However, the lipid mixture of the nuclear envelope/endoplasmic reticulum possesses a relatively low amount of cholesterol and sphingomyelin compared with the plasma membrane (30) and does not form detectable lipid rafts (40). Accordingly, we did not observe phase separation between a cholesterol/sphingomyelin-enriched ordered and a disordered phase when pUL31 was added to pUL34-GUVs (Fig. 3C). Therefore, there is no indication for a locally induced phase separation, giving rise to the observed invaginations. In addition, changes in lipid composition, especially the localized generation of non-cone shaped lipids or their enrichment, such as the unconventional phospholipid lysobisphosphatidic acid on internal vesicles of multivesicular bodies (59), can contribute to membrane curving. However, there is no evidence that pUL31 mediates lipid modifying reactions or has a binding preference for a specific lipid.
Therefore, we prefer a model in which pUL31 acts as a scaffolding protein self-assembling at the inner surface of the forming vesicle. pUL31 is monomeric and does not detectably self-interact in solution (Fig. 4, B–D). Artificial membrane tethering of limiting amounts of pUL31 that are not sufficient to promote membrane invaginations can recruit soluble pUL31 (Fig. 4A), which, in turn, induces intralumenal vesicle formation. This effect is caused by membrane-induced pUL31 oligomerization, as suggested by confocal and atomic force microscopy on supported lipid bilayers. Consistently, budding sites at the GUV membranes show local enrichment of pUL34-pUL31 or pUL31, respectively (Figs. 1I and 2E). Although we cannot exclude the contribution of other proteins, in cells, budding sites at the inner nuclear membrane are more electron-dense, an observation that implies high protein density (8, 10). Interestingly, a recent study shows that a pUL31-pUL34 complex lacking the pUL34 transmembrane domain forms ordered assemblies when recruited to membranes (21). Our study suggests that pUL31 alone can oligomerize on membranes and induce vesicle formation. For the Kaposi sarcoma-associated herpesvirus pUL31 homolog ORF69, it has been suggested that it is required for membrane remodelling into circular virion-size vesicles, whereas the pUL34 homolog ORF67 results in membrane proliferation (60). A double point mutation in HSV-1 pUL34 (D35A/E37A) causes a blockage in nuclear envelope vesicle formation without preventing pUL31 membrane recruitment (61). When tethered to GUVs, the corresponding pUL31/pUL34 fails to induce membrane deformation (21). The defect of pUL34 (D35A/E37A) during infection could be overcome by a mutation in pUL31 that restored the ability to induce membrane deformation (61). This indicates that pUL34, in addition to pUL31 recruitment, plays a supportive and regulatory role in the induction of vesicles (62). It is possible that pUL34 undergoes a conformational change upon pUL31 binding that either brings the protein closer to the membrane or triggers multimerization of the complex, features that might be blocked in the D35A/E37A mutation.
The vesicle budding at the nuclear envelope is reminiscent of the invagination processes mediated by the ESCRT machinery during formation of intraendosomal vesicles in multivesicular body formation. In this reaction pathway, ESCRT-I and ESCRT-II complexes direct membrane budding away from the cytosol, and ESCRT-III cleaves the bud necks from their cytosolic faces (23, 63, 64). In a mechanistically related process during HIV egress at the plasma membrane, the viral gag protein assembles and drives initial bud formation but requires the ESCRT complex components for membrane scission (65, 66). Gag protein assembly on the cytoplasmic side of the plasma membrane is thought to be the driving force for bud formation in HIV, and similar roles for membrane-associated proteins are implicated in rhabdovirus, filovirus, arenavirus, and paramyxovirus formation (37). In contrast to the gag/ESCRT machinery, which mediates HIV release at the plasma membrane (67), the nuclear egress complex requires no energy input for both membrane budding and scission,3 although we cannot exclude that, in the cellular situation, an energy-dependent step is involved. Interestingly, although, in the case of gag/ESCRT-mediated vesicle formation, several proteins are required and the steps of membrane budding and scission are separated functionally, a single protein, pUL31, can execute this task in herpesvirus nuclear egress. This suggests that pUL31 has the intrinsic ability to oligomerize on the membrane and cleave its own bud neck. How this is achieved mechanistically is an interesting avenue for future research. In contrast to a previous report, we did not detect a regular structural pattern on the membrane surface (Fig. 5), most likely because of the absence of pUL34. This suggest that pUL34 is, in addition to pUL31 recruitment, required for formation of the hexagonal arrangement and drives the otherwise unstructured oligomerization of pUL31 into an ordered pattern.
Although we cannot exclude the presence of smaller-sized vesicles inside the GUVs because of the refraction limit of light microscopy and the rapid vesicle movement, the vesicles observed in the in vitro system are considerably larger than the vesicles formed in the nuclear envelope lumen during viral egress (68) but also in the absence of capsids when overexpressing pUL31 and pUL34 (10, 20). Notably, a similar increase in vesicle diameters has been observed when studying budding and/or vesicle formation on GUVs mediated by the ESCRT machinery or other viral proteins, including pUL31/pUL34 (21, 63, 69–71). A number of factors could, in the case of our study, account for the size difference. The inner nuclear membrane is tightly connected to the underlying lamina and the chromatin by integral membrane proteins (1, 2). This could restrain lateral mobility of the membrane and, consequently, impact the size of the vesicles. Moreover, although pUL31 can, on its own, promote vesicle budding and scission, it is conceivable that cellular (and additional viral) factors contribute to the process in vivo, which could restrict vesicle size. The fact that a similar vesicle-mediated nuclear export of large ribonucleoprotein particles bypassing nuclear pore complexes is found (24) makes it likely that a cellular machinery exists that mediates membrane budding and scission at the inner nuclear membrane as well as fusion of these vesicles with the outer nuclear membrane and that could participate in herpesvirus nuclear egress.
In summary, our work establishes that pUL34-mediated membrane recruitment of pUL31 drives vesicle budding and constriction in a biochemically well defined system. It shows that a single protein, pUL31, is sufficient to induce inwardly directed membrane deformation and scission. It will be interesting to see whether cellular orthologs of herpesvirus pUL31 exist, especially for the process of RNP egress at the nuclear envelope, which is topologically comparable with herpesvirus nuclear egress. Finally, the methods established and employed here to express and reconstitute pUL34 into GUVs are generally applicable for other single and multiple transmembrane-spanning proteins and, therefore, present a valuable tool for studying these proteins in minimal and well defined membrane systems.
Acknowledgments
We thank C. Liebig (Imaging Facility MPI DevBiol) for help with confocal microscopy and image analysis; V. Betaneli and N. Eisenhardt for the introduction to GUV preparation; V. Ahl for help with the MALLS experiment; C. Sieverding and S. Astrinidis for technical support; and N. Eisenhardt, D. Moreno, K. Schellhaus, and A. Schooley for critical reading of the manuscript.
This work was supported by funding from the Max Planck Society (to W. A.) and by German Research Foundation Grant DFG Me 854/12-1.
M. Lorenz, B. Vollmer, J. D. Unsay, B. G. Klupp, A. J. García-Sáez, T. C. Mettenleiter, and W. Antonin, unpublished observations.
- GUV
- giant unilamellar vesicle
- DiDC18
- 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine,4-chlorobenzenesulfonate salt
- EGFP
- enhanced GFP
- Ni-NTA
- nickel-nitrilotriacetic acid
- DGS
- 1,2-dioleoyl-sn-glycero-3-(N-(5-amino-1-carboxypentyl)iminodiacetic acid) succinyl
- ILV
- intralumenal vesicle
- COP
- coat protein complex
- ESCRT
- endosomal sorting complex required for transport
- TEV
- tobacco etch virus.
REFERENCES
- 1. Hetzer M. W., Wente S. R. (2009) Border control at the nucleus: biogenesis and organization of the nuclear membrane and pore complexes. Dev. Cell 17, 606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Schooley A., Vollmer B., Antonin W. (2012) Building a nuclear envelope at the end of mitosis: coordinating membrane reorganization, nuclear pore complex assembly, and chromatin de-condensation. Chromosoma 121, 539–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Adams R. L., Wente S. R. (2013) Uncovering nuclear pore complexity with innovation. Cell 152, 1218–1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kobiler O., Drayman N., Butin-Israeli V., Oppenheim A. (2012) Virus strategies for passing the nuclear envelope barrier. Nucleus 3, 526–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cohen S., Au S., Panté N. (2011) How viruses access the nucleus. Biochim. Biophys. Acta 1813, 1634–1645 [DOI] [PubMed] [Google Scholar]
- 6. Skepper J. N., Whiteley A., Browne H., Minson A. (2001) Herpes simplex virus nucleocapsids mature to progeny virions by an envelopment → deenvelopment → reenvelopment pathway. J. Virol. 75, 5697–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson D. C., Baines J. D. (2011) Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9, 382–394 [DOI] [PubMed] [Google Scholar]
- 8. Mettenleiter T. C., Müller F., Granzow H., Klupp B. G. (2013) The way out: what we know and do not know about herpesvirus nuclear egress. Cell. Microbiol. 15, 170–178 [DOI] [PubMed] [Google Scholar]
- 9. Fuchs W., Klupp B. G., Granzow H., Osterrieder N., Mettenleiter T. C. (2002) The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76, 364–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Klupp B. G., Granzow H., Fuchs W., Keil G. M., Finke S., Mettenleiter T. C. (2007) Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 7241–7246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lake C. M., Hutt-Fletcher L. M. (2004) The Epstein-Barr virus BFRF1 and BFLF2 proteins interact and coexpression alters their cellular localization. Virology 320, 99–106 [DOI] [PubMed] [Google Scholar]
- 12. Reynolds A. E., Ryckman B. J., Baines J. D., Zhou Y., Liang L., Roller R. J. (2001) U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75, 8803–8817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schnee M., Ruzsics Z., Bubeck A., Koszinowski U. H. (2006) Common and specific properties of herpesvirus UL34/UL31 protein family members revealed by protein complementation assay. J. Virol. 80, 11658–11666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sam M. D., Evans B. T., Coen D. M., Hogle J. M. (2009) Biochemical, biophysical, and mutational analyses of subunit interactions of the human cytomegalovirus nuclear egress complex. J. Virol. 83, 2996–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Milbradt J., Auerochs S., Sticht H., Marschall M. (2009) Cytomegaloviral proteins that associate with the nuclear lamina: components of a postulated nuclear egress complex. J. Gen. Virol. 90, 579–590 [DOI] [PubMed] [Google Scholar]
- 16. Reynolds A. E., Liang L., Baines J. D. (2004) Conformational changes in the nuclear lamina induced by herpes simplex virus type 1 require genes U(L)31 and U(L)34. J. Virol. 78, 5564–5575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bjerke S. L., Roller R. J. (2006) Roles for herpes simplex virus type 1 UL34 and US3 proteins in disrupting the nuclear lamina during herpes simplex virus type 1 egress. Virology 347, 261–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mou F., Forest T., Baines J. D. (2007) US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J. Virol. 81, 6459–6470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Muranyi W., Haas J., Wagner M., Krohne G., Koszinowski U. H. (2002) Cytomegalovirus recruitment of cellular kinases to dissolve the nuclear lamina. Science 297, 854–857 [DOI] [PubMed] [Google Scholar]
- 20. Desai P. J., Pryce E. N., Henson B. W., Luitweiler E. M., Cothran J. (2012) Reconstitution of the Kaposi's sarcoma-associated herpesvirus nuclear egress complex and formation of nuclear membrane vesicles by coexpression of ORF67 and ORF69 gene products. J. Virol. 86, 594–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bigalke J. M., Heuser T., Nicastro D., Heldwein E. E. (2014) Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat. Commun. 5, 4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kirchhausen T. (2000) Three ways to make a vesicle. Nat. Rev. Mol. Cell Biol. 1, 187–198 [DOI] [PubMed] [Google Scholar]
- 23. Hurley J. H., Hanson P. I. (2010) Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nat. Rev. Mol. Cell Biol. 11, 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Speese S. D., Ashley J., Jokhi V., Nunnari J., Barria R., Li Y., Ataman B., Koon A., Chang Y. T., Li Q., Moore M. J., Budnik V. (2012) Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149, 832–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roosild T. P., Greenwald J., Vega M., Castronovo S., Riek R., Choe S. (2005) NMR structure of Mistic, a membrane-integrating protein for membrane protein expression. Science 307, 1317–1321 [DOI] [PubMed] [Google Scholar]
- 26. Theerthagiri G., Eisenhardt N., Schwarz H., Antonin W. (2010) The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J. Cell Biol. 189, 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Eisenhardt N., Redolfi J., Antonin W. (2014) Interaction of Nup53 with Ndc1 and Nup155 is required for nuclear pore complex assembly. J. Cell Sci. 127, 908–921 [DOI] [PubMed] [Google Scholar]
- 28. Angelova M. I., Dimitrov D. S. (1986) Liposome electroformation. Faraday Discuss. 81, 303–311 [Google Scholar]
- 29. Unsay J. D., Cosentino K., Subburaj Y., García-Sáez A. J. (2013) Cardiolipin effects on membrane structure and dynamics. Langmuir 29, 15878–15887 [DOI] [PubMed] [Google Scholar]
- 30. Kleinig H. (1970) Nuclear membranes from mammalian liver: II: lipid composition. J. Cell Biol. 46, 396–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vollmer B., Schooley A., Sachdev R., Eisenhardt N., Schneider A. M., Sieverding C., Madlung J., Gerken U., Macek B., Antonin W. (2012) Dimerization and direct membrane interaction of Nup53 contribute to nuclear pore complex assembly. EMBO J. 31, 4072–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ulbert S., Platani M., Boue S., Mattaj I. W. (2006) Direct membrane protein-DNA interactions required early in nuclear envelope assembly. J. Cell Biol. 173, 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Voeltz G. K., Prinz W. A., Shibata Y., Rist J. M., Rapoport T. A. (2006) A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586 [DOI] [PubMed] [Google Scholar]
- 34. Walser P. J., Ariotti N., Howes M., Ferguson C., Webb R., Schwudke D., Leneva N., Cho K. J., Cooper L., Rae J., Floetenmeyer M., Oorschot V. M., Skoglund U., Simons K., Hancock J. F., Parton R. G. (2012) Constitutive formation of caveolae in a bacterium. Cell 150, 752–763 [DOI] [PubMed] [Google Scholar]
- 35. McMahon H. T., Gallop J. L. (2005) Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438, 590–596 [DOI] [PubMed] [Google Scholar]
- 36. Garoff H., Simons K. (1974) Location of the spike glycoproteins in the Semliki Forest virus membrane. Proc. Natl. Acad. Sci. U.S.A. 71, 3988–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Welsch S., Müller B., Kräusslich H. G. (2007) More than one door: budding of enveloped viruses through cellular membranes. FEBS Lett. 581, 2089–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schuster F., Klupp B. G., Granzow H., Mettenleiter T. C. (2012) Structural determinants for nuclear envelope localization and function of pseudorabies virus pUL34. J. Virol. 86, 2079–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lingwood D., Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50 [DOI] [PubMed] [Google Scholar]
- 40. van Meer G., Voelker D. R., Feigenson G. W. (2008) Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9, 112–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mettenleiter T. C. (2002) Herpesvirus assembly and egress. J. Virol. 76, 1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jokhi V., Ashley J., Nunnari J., Noma A., Ito N., Wakabayashi-Ito N., Moore M. J., Budnik V. (2013) Torsin mediates primary envelopment of large ribonucleoprotein granules at the nuclear envelope. Cell Rep. 3, 988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Klupp B. G., Granzow H., Mettenleiter T. C. (2011) Nuclear envelope breakdown can substitute for primary envelopment-mediated nuclear egress of herpesviruses. J. Virol. 85, 8285–8292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chang Y. E., Van Sant C., Krug P. W., Sears A. E., Roizman B. (1997) The null mutant of the U(L)31 gene of herpes simplex virus 1: construction and phenotype in infected cells. J. Virol. 71, 8307–8315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Granato M., Feederle R., Farina A., Gonnella R., Santarelli R., Hub B., Faggioni A., Delecluse H. J. (2008) Deletion of Epstein-Barr virus BFLF2 leads to impaired viral DNA packaging and primary egress as well as to the production of defective viral particles. J. Virol. 82, 4042–4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Haugo A. C., Szpara M. L., Parsons L., Enquist L. W., Roller R. J. (2011) Herpes simplex virus 1 pUL34 plays a critical role in cell-to-cell spread of virus in addition to its role in virus replication. J. Virol. 85, 7203–7215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ledesma M. D., Dotti C. G. (2003) Membrane and cytoskeleton dynamics during axonal elongation and stabilization. Int. Rev. Cytol. 227, 183–219 [DOI] [PubMed] [Google Scholar]
- 48. Merrifield C. J. (2004) Seeing is believing: imaging actin dynamics at single sites of endocytosis. Trends Cell Biol. 14, 352–358 [DOI] [PubMed] [Google Scholar]
- 49. Vennema H., Godeke G. J., Rossen J. W., Voorhout W. F., Horzinek M. C., Opstelten D. J., Rottier P. J. (1996) Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 15, 2020–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Farsad K., Ringstad N., Takei K., Floyd S. R., Rose K., De Camilli P. (2001) Generation of high curvature membranes mediated by direct endophilin bilayer interactions. J. Cell Biol. 155, 193–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Boucrot E., Pick A., Çamdere G., Liska N., Evergren E., McMahon H. T., Kozlov M. M. (2012) Membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell 149, 124–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Stachowiak J. C., Schmid E. M., Ryan C. J., Ann H. S., Sasaki D. Y., Sherman M. B., Geissler P. L., Fletcher D. A., Hayden C. C. (2012) Membrane bending by protein-protein crowding. Nat. Cell Biol. 14, 944–949 [DOI] [PubMed] [Google Scholar]
- 53. Roux A., Cuvelier D., Nassoy P., Prost J., Bassereau P., Goud B. (2005) Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J. 24, 1537–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Baumgart T., Hess S. T., Webb W. W. (2003) Imaging coexisting fluid domains in biomembrane models coupling curvature and line tension. Nature 425, 821–824 [DOI] [PubMed] [Google Scholar]
- 55. Bacia K., Schwille P., Kurzchalia T. (2005) Sterol structure determines the separation of phases and the curvature of the liquid-ordered phase in model membranes. Proc. Natl. Acad. Sci. U.S.A. 102, 3272–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brügger B., Glass B., Haberkant P., Leibrecht I., Wieland F. T., Kräusslich H. G. (2006) The HIV lipidome: a raft with an unusual composition. Proc. Natl. Acad. Sci. U.S.A. 103, 2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Aloia R. C., Jensen F. C., Curtain C. C., Mobley P. W., Gordon L. M. (1988) Lipid composition and fluidity of the human immunodeficiency virus. Proc. Natl. Acad. Sci. U.S.A. 85, 900–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ono A., Freed E. O. (2005) Role of lipid rafts in virus replication. Adv. Virus Res. 64, 311–358 [DOI] [PubMed] [Google Scholar]
- 59. Matsuo H., Chevallier J., Mayran N., Le Blanc I., Ferguson C., Fauré J., Blanc N. S., Matile S., Dubochet J., Sadoul R., Parton R. G., Vilbois F., Gruenberg J. (2004) Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science 303, 531–534 [DOI] [PubMed] [Google Scholar]
- 60. Luitweiler E. M., Henson B. W., Pryce E. N., Patel V., Coombs G., McCaffery J. M., Desai P. J. (2013) Interactions of the Kaposi's sarcoma-associated herpesvirus nuclear egress complex: ORF69 is a potent factor for remodeling cellular membranes. J. Virol. 87, 3915–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Roller R. J., Bjerke S. L., Haugo A. C., Hanson S. (2010) Analysis of a charge cluster mutation of herpes simplex virus type 1 UL34 and its extragenic suppressor suggests a novel interaction between pUL34 and pUL31 that is necessary for membrane curvature around capsids. J. Virol. 84, 3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Roller R. J., Haugo A. C., Kopping N. J. (2011) Intragenic and extragenic suppression of a mutation in herpes simplex virus 1 UL34 that affects both nuclear envelope targeting and membrane budding. J. Virol. 85, 11615–11625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wollert T., Hurley J. H. (2010) Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464, 864–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wollert T., Wunder C., Lippincott-Schwartz J., Hurley J. H. (2009) Membrane scission by the ESCRT-III complex. Nature 458, 172–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sundquist W. I., Kräusslich H. G. (2012) HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2, a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Van Engelenburg S. B., Shtengel G., Sengupta P., Waki K., Jarnik M., Ablan S. D., Freed E. O., Hess H. F., Lippincott-Schwartz J. (2014) Distribution of ESCRT machinery at HIV assembly sites reveals virus scaffolding of ESCRT subunits. Science 343, 653–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Baumgärtel V., Ivanchenko S., Dupont A., Sergeev M., Wiseman P. W., Kräusslich H. G., Bräuchle C., Müller B., Lamb D. C. (2011) Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat. Cell Biol. 13, 469–474 [DOI] [PubMed] [Google Scholar]
- 68. Mettenleiter T. C., Klupp B. G., Granzow H. (2009) Herpesvirus assembly: an update. Virus Res. 143, 222–234 [DOI] [PubMed] [Google Scholar]
- 69. Shnyrova A. V., Ayllon J., Mikhalyov I. I., Villar E., Zimmerberg J., Frolov V. A. (2007) Vesicle formation by self-assembly of membrane-bound matrix proteins into a fluidlike budding domain. J. Cell Biol. 179, 627–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Solon J., Gareil O., Bassereau P., Gaudin Y. (2005) Membrane deformations induced by the matrix protein of vesicular stomatitis virus in a minimal system. J. Gen. Virol. 86, 3357–3363 [DOI] [PubMed] [Google Scholar]
- 71. Rossman J. S., Jing X., Leser G. P., Lamb R. A. (2010) Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 142, 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]