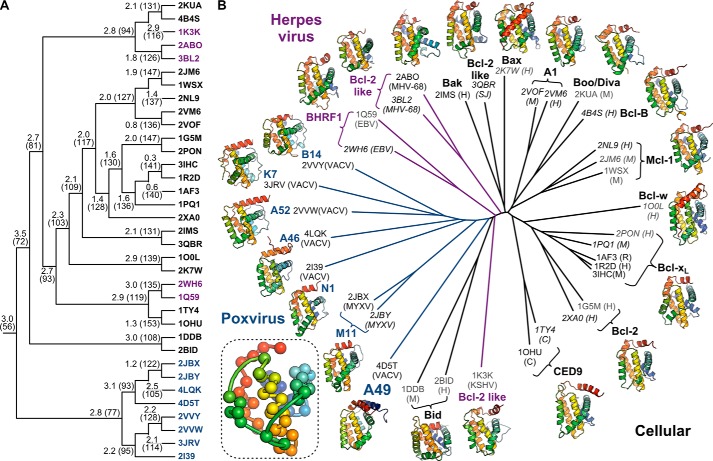
FIGURE 4.
Poxvirus Bcl-2 proteins are structurally closer to each other than to other cellular or viral Bcl-2 proteins. A, clustering of cellular and viral Bcl-2 proteins to determine the minimal structural “core” of the Bcl-2 fold. PDB identifiers of structures used are shown as leaves on the tree, colored by source organism (black, cellular; magenta, herpesvirus; blue, poxvirus). Cα atom root mean square deviation (Å) and number of equivalent Cα atoms, which corresponds to the size of the common core, are shown at each branch point. For higher branches in the tree, superpositions were performed between these common cores to iteratively determine the minimal core of the Bcl-2 fold. B, structure-based phylogenetic tree showing the relationship between cellular, herpesvirus, and poxvirus Bcl-2 proteins. PDB codes for each structure used are given with their species of origin in parentheses (human (H), mouse (M), rat (R), Caenorhabditis elegans (C), Schistosoma japonicum (SJ), vaccinia virus (VACV), myxoma virus (MYXV), Kaposi sarcoma herpesvirus (KSHV), murine γ-herpesvirus 68 (MHV-68), and Epstein-Barr virus (EBV)). Structures determined by x-ray crystallography are labeled in black, NMR models are shown in gray, and structures determined with BH3 peptides or C-terminal tails bound in the surface groove are italicized. Ribbon diagrams of representative structures for each protein are shown, color-ramped from blue (N terminus) to red (C terminus). The inset shows the 56 Cα atoms that comprise the minimal core of the Bcl-2 fold, colored as above. Coordinate files containing the core Cα atoms of the Bcl-2 fold plus superpositions of the 35 representative Bcl-2 family structures upon this core are supplied as supplemental material.