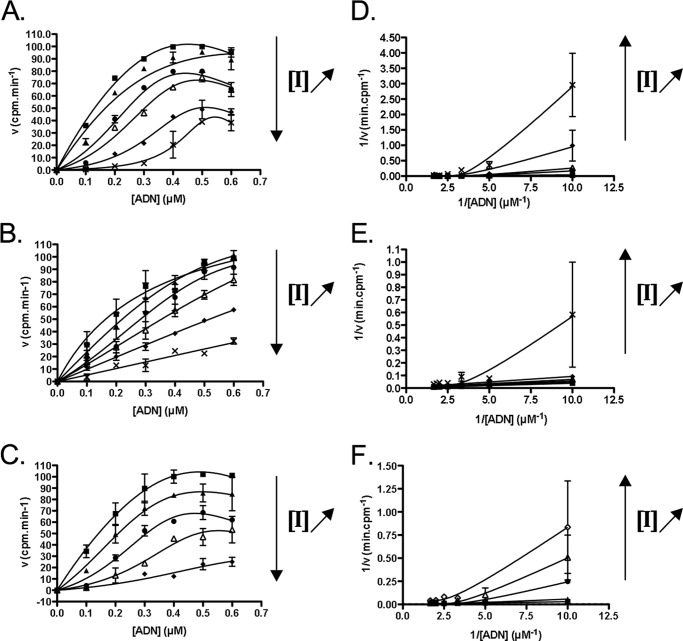
FIGURE 2.
DNA-competition studies with SGI-1027 and its analogs in a DNMT1 enzymatic assay. A–C, initial velocity plots against DNA concentrations for different concentrations of SGI-1027 (A), 5 (B), and 31 (C). Squares represent points in the absence of the inhibitor. For SGI-1027 (A): triangles, circles, empty triangles, diamonds, and crosses represent 5.6, 10, 14.7, 21.5, and 32 μm, respectively. For 5 (B): triangles, circles, empty triangles, diamonds, and crosses represent 3.2, 4.2, 5.6, 7.5, and 10 μm, respectively. For 31 (C): triangles, circles, empty triangles, and diamonds represent 10, 25, 40, and 55 μm, respectively. Plain lines represent fittings with the Copeland and Horiuchi (17) competitive model. D–F, Lineweaver-Burk (or double reciprocal) plots for SGI-1027 (D), 5 (E), and 31 (F). For the corresponding concentrations, the same legend is applied as for A–C. Plain lines represent fittings with the double-reciprocal of Copeland and Horiuchi (17) competitive model. The means of two experiments are displayed with the corresponding S.E.