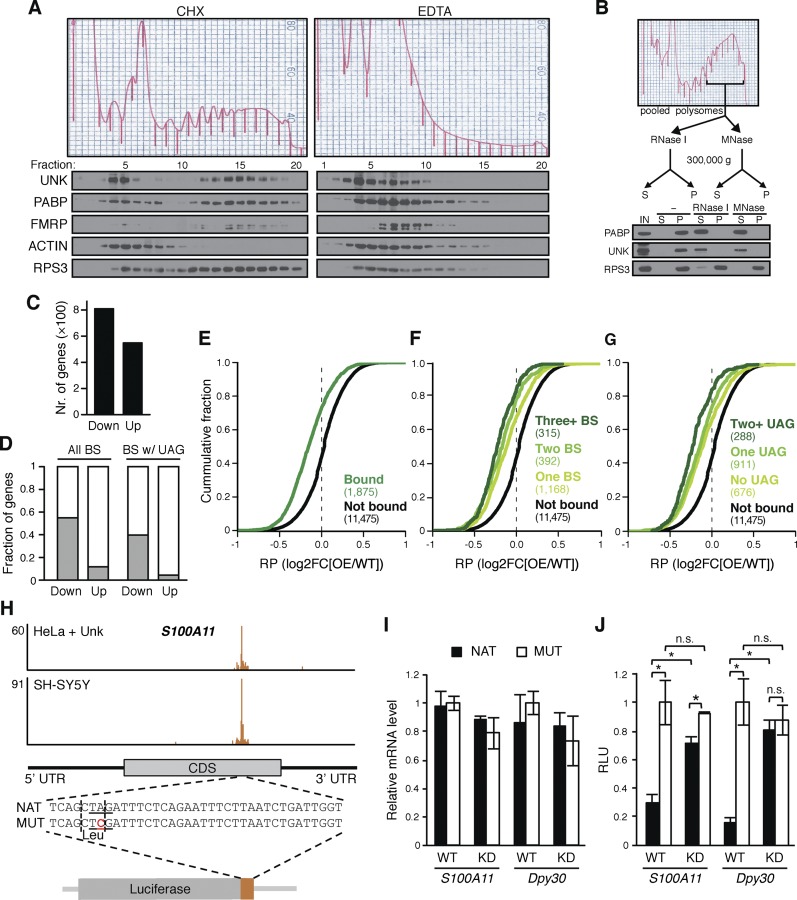
Figure 6.
Unkempt represses translation of its target messages. (A,B) RNA-dependent association of Unkempt with polyribosomes. (A) Polysome profiling of SH-SY5Y cells harvested in the presence of cycloheximide (CHX) or EDTA and immunoblot analysis of individual fractions for the indicated proteins. See also Supplemental Figure S8A. (B) Digestion of polyribosomal fractions with RNase I or MNase followed by immunoblot analysis of the indicated proteins for their release into the supernatant (S) from the pelleted (P) fraction. (IN) Input. (C) Ribosome profiling data showing total numbers of genes with significantly decreased (Down) or increased (Up) RPF count in Dox-treated GFP and Unkempt-inducible versus GFP-only-inducible HeLa cells (false discovery rate [FDR] < 5%). (D) Fractions of genes with altered RPF counts as shown in C that are bound by Unkempt, considering all binding sites (All BS) or binding sites harboring the UAG motif (BS w/UAG). (E–G) Cumulative distributions of changes in ribosome occupancy for all transcripts containing Unkempt-binding sites (Bound); transcripts with one (One BS), two (Two BS), or three or more (Three+ BS) binding sites; transcripts without the UAG motif (No UAG), with one UAG motif (One UAG), or with two or more UAG motifs (Two+ UAG) within the binding sites; and control transcripts lacking Unkempt-binding sites (Not bound). The number of genes in each category is indicated in parentheses. Comparison of either set of Unkempt target transcripts with the nonbound controls showed a significant difference (P < 0.0001). (H–J) Translational repression by Unkempt requires the UAG motif. (H) Extension of the luciferase gene with native (NAT) or point-mutated (MUT) sequence corresponding to the Unkempt-binding site within the human S100A11 transcript. The mutation in the critical UAG motif (underlined) preserves the amino acid (Leu) encoded by the affected codon (vertical dashed lines). (I,J) Dual-luciferase assay using the native or point-mutated hybrid luciferase in wild-type (WT) and Unkempt knockdown (KD) SH-SY5Y cells (see also Supplemental Fig. S9C). Relative levels of the hybrid luciferase transcripts (I) and relative luminescence units (RLU) (J) are shown. Error bars indicate SD (n = 3). (*) P < 0.05, Student’s t-test.