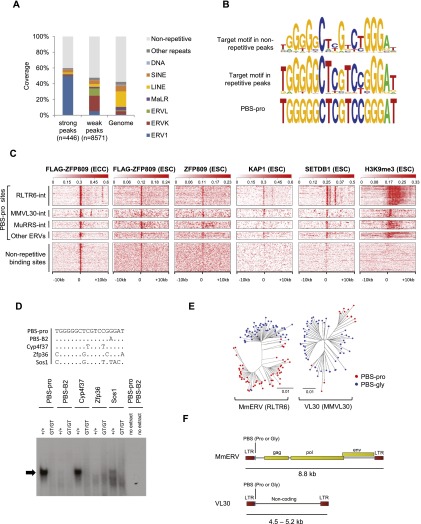
Figure 1.
Genome-wide mapping of ZFP809-binding sites. (A) Repeat content of strong (enrichment >50) and weak (enrichment <50) Flag-ZFP809 ChIP-seq peaks and the mouse genome as determined by genomic overlap with University of California at Santa Cruz (UCSC) RepeatMasker annotations. (B) Consensus Flag-ZFP809 target motif derived from the 100 top-scored nonrepetitive peaks (top) and the corresponding target consensus in strong repetitive peaks (middle). The PBS-pro sequence is shown at the bottom. (C) Heat maps of ChIP-seq data at ERV-associated PBS-pro sites and strong nonrepetitive Flag-ZFP809 ChIP-seq peak regions. Ten-kilobase regions are displayed with the PBS-pro sequence or nonrepetitive peaks located at the center. PBS-pro sites were grouped according to flanking ERV sequences and are orientated 5′ to 3′. (D) EMSA with nuclear extracts from wild-type and Zfp809 mutant (Zfp809GT/GT) ESCs. Sequences of dsDNA probes (+ strand, 5′ to 3′) are shown in the top panel. The repressor complex binding to the PBS-pro probe is indicated by an arrow. (E) Neighbor-joining phylogenetic trees of genomic MmERV and VL30 elements containing PBS-pro or PBS-gly. The pol gene or the noncoding internal region was used to create trees for MmERV and VL30 elements, respectively. (F) Schematic representation of MmERV and VL30 consensus sequences. Yellow rectangles indicate coding regions for retroviral proteins.