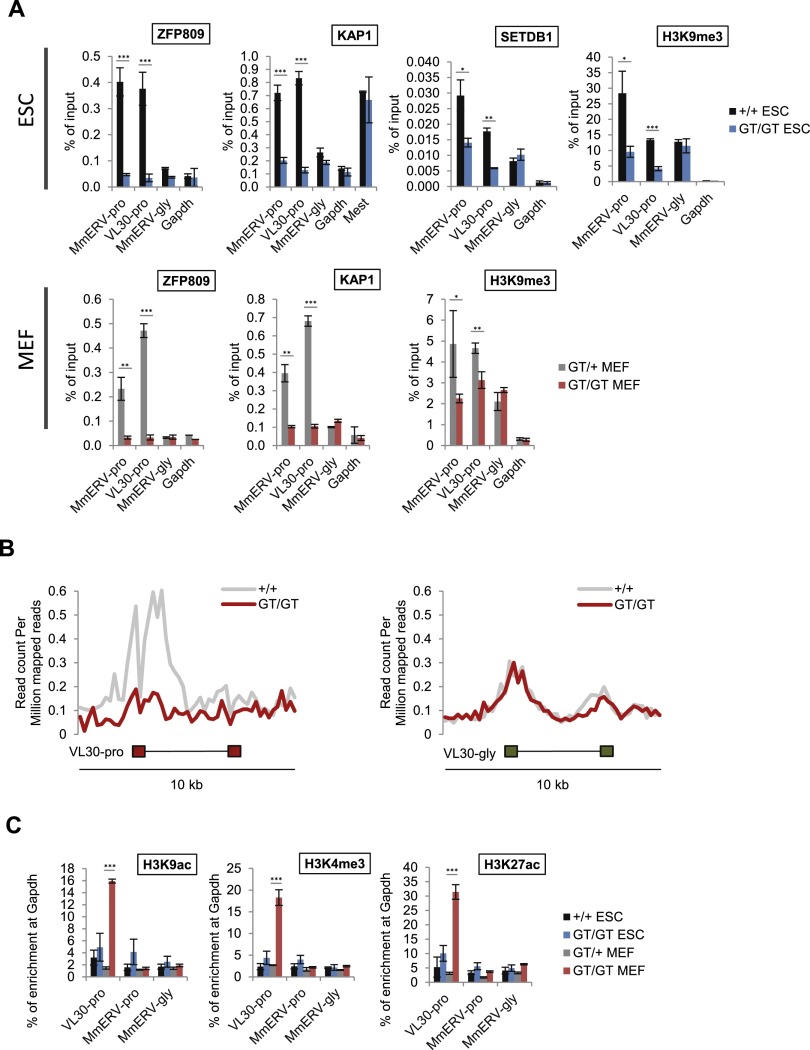
Figure 3.
ZFP809 is required to recruit KAP1/SETDB1 to ERVs and establish heterochromatin marks. (A) ChIP-qPCR (ZFP809, KAP1, and SETDB1) or native ChIP (NChIP)-qPCR (H3K9me3) analysis of ERVs in Zfp809GT ESCs (top panel) and MEFs (bottom panel). Enrichment is shown as mean percentage of input DNA (n = 3 technical replicates). Error bars represent SD. P-values (by unpaired t-tests) are indicated as follows: (***) P < 0.0005; (**) 0.0005<P < 0.005; (*) 0.005<P < 0.05. (B) NGS plots showing average H3K9me3 NChIP-seq read densities at 10-kb regions around genomic VL30-pro (n = 15) and VL30-gly (n = 65) elements in Zfp809+/+ and Zfp809GT/GT ESCs. (C) ChIP-qPCR analysis of markers of active chromatin in Zfp809GT MEFs and ESCs. Relative enrichment was normalized to input DNA and is shown as mean percentage (n = 3 technical replicates) of enrichment at the Gapdh promoter. Error bars represent SD. P-values (by unpaired t-tests) are indicated as described above.