Figure 1.
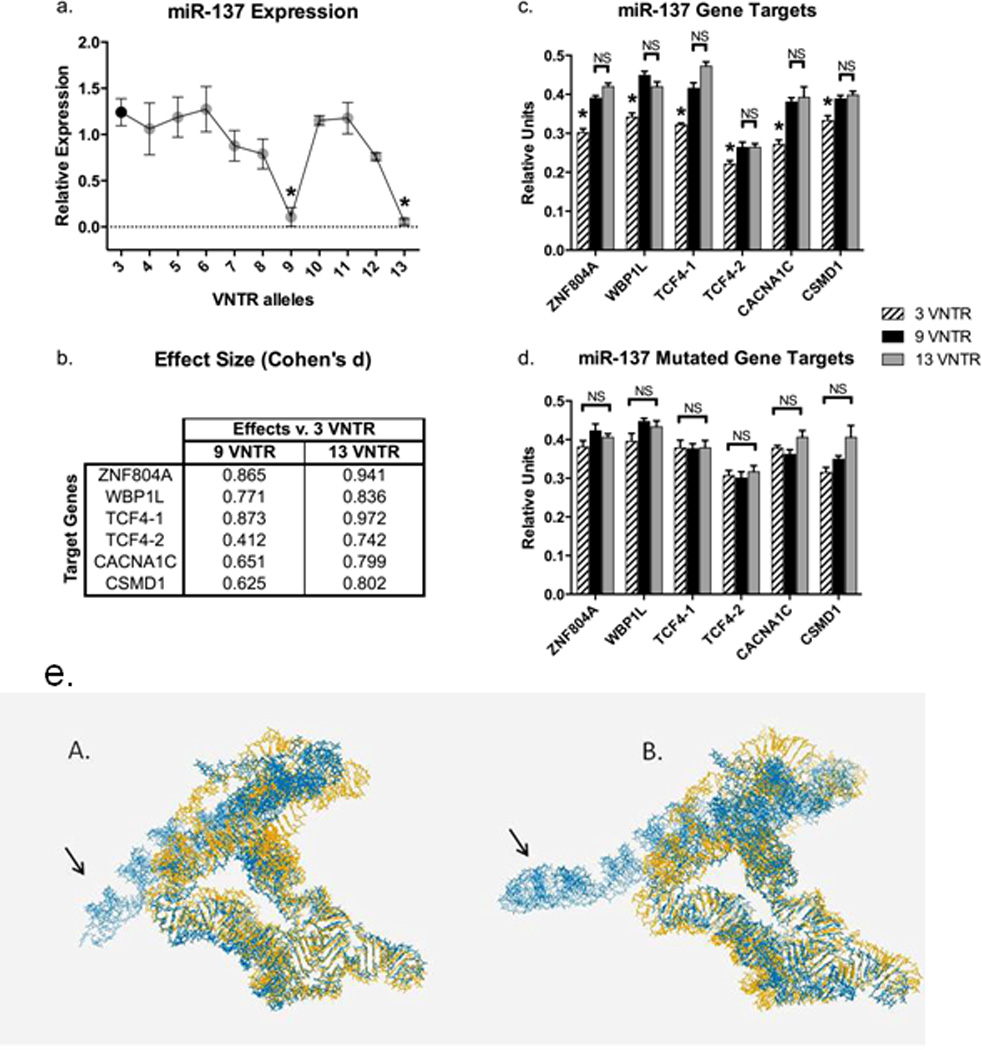
(a) MiRNA expression was assessed by qPCR and relative expression values calculated using 2−ΔΔCt. Data were analyzed using the Kruskal-Wallis test, followed by Dunn’s post-hoc test correcting for multiple testing (GraphPad Prism 6). Two VNTR alleles, 9 and 13, exhibited significantly reduced expression of miR-137, while 7, 8, and 12 showed a trend of reduction in miR-137 expression.
Asterisks indicate significant differences. All experiments were performed in triplicate and results shown are means ± SEM.
(b) Effect sizes of luciferase activity differences (Cohen’s d) between 9 and 13 VNTR alleles compared to the 3 VNTR allele.
(c) Luciferase assay in HEK293 cells; co-expression of miR-137-VNTR alleles with wild-type gene target sequences of schizophrenia candidate genes: ZNF804A, WBP1L, TCF4-1, TCF4-2, CACNA1C, and CSMD1. 9 and 13 VNTR alleles exhibited significantly higher luciferase activity compared to 3 VNTR allele for all target genes. (d) Once gene target sequences were mutated no difference between 9 and 13 compared to 3 was observed. The data were analyzed using Mann-Whitney test followed by Bonferroni correction for multiple testing. Asterisks indicate significant differences. NS indicates no significant differences. All experiments were performed in triplicate and results shown are means ± SEM.
(e) The impact of the VNTR on the 3D structure of the miR-137 precursor was analyzed using RNAcomposer (http://rnacomposer.cs.put.poznan.pl/). The total length of the miR-137 precursor sequences varied from 350 bases (3 VNTR allele) to 500 bases (13 VNTR allele). In this composite figure, the 3D structure of miR-137 for the 3 VNTR allele is colored in yellow, whereas the 9 (panel A) and 13 (panel B) VNTR alleles are overlaid in blue. Although, the 3D structures share similarity, the local substructures containing the 9 and 13 VNTR alleles deviate significantly from the folding pattern of the 3 VNTR allele (noted by the arrows).
