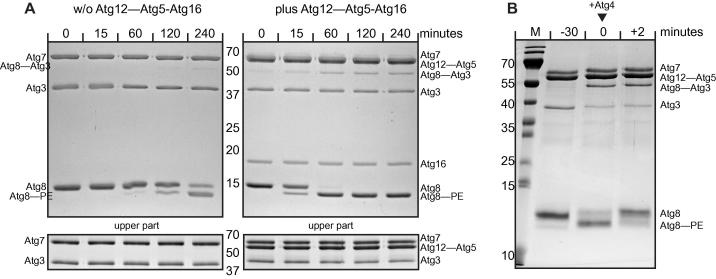
Fig. 3.
In vitro Atg8 lipidation/de-lipidation reaction using SUVs. (A) Two Coomassie-stained gels of two time series of Atg8 conjugation reactions using SUVs derived from the E. coli lipids. The first reaction was conducted in the absence (left) and the second reaction was conducted in the presence (right) of the Atg12–Atg5-Atg16 complex. The lower gels shown in the bottom were run at lower polyacrylamide concentrations (12% as opposed to 16% for the upper two gels) in order to separate the Atg7 protein from the Atg12–Atg5 conjugate. Note the massive stimulation of Atg8 lipidation in the presence of the Atg12–Atg5-Atg16 complex. The numbers between the gels indicate the molecular weights of the marker bands in kDa. (B) Coomassie-stained gel of an Atg8 lipidation/de-lipidation reaction. The SUVs were composed of 40% POPC, 35% POPS, 20% POPE and 5% PI3P. Atg8 was first completely lipidated in the presence of Atg3, Atg7, the Atg12–Atg5 conjugate and ATP and MgCl2. After 30 min Atg4 and EDTA were added to final concentrations of 0.07 μM and 2 mM, respectively. After 2 min Atg8 was completely de-lipidated. The numbers to the left of the gel indicate the molecular weights of the marker bands in kDa. Note that the 35 kDa and 15 kDa bands separate into two bands in the presence of 6 M urea. (M: marker).