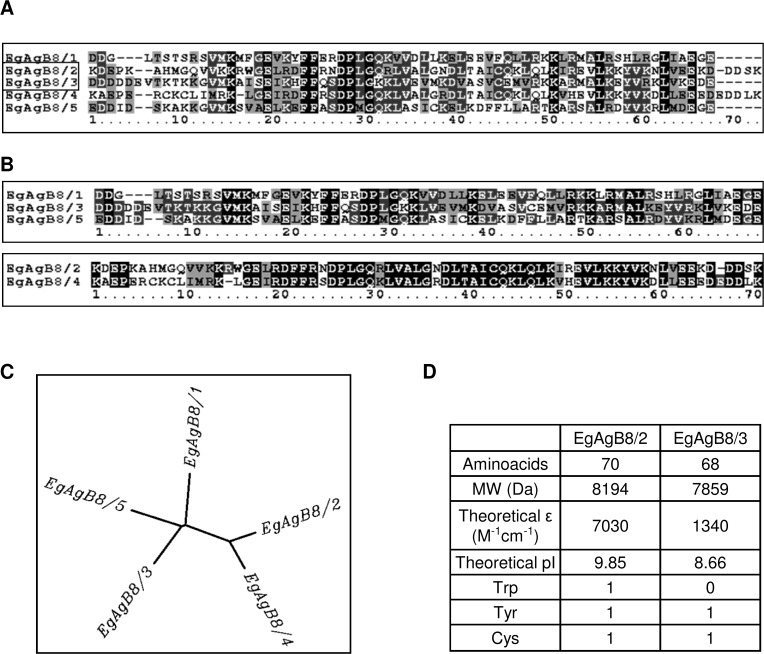
Fig 1. Amino acid sequence analysis of EgAgB8 subunits.
Analysis of EgAgB8 subunits were undertaken using Biology Workbench 3.2 (University of Illinois, National Center for Supercomputing Applications, USA). (A) Alignment of amino acid sequences of the mature peptides of EgAgB8/1, EgAgB8/2, EgAgB8/3, EgAgB8/4 and EgAgB8/5, using ClustalW from Biology Workbench 3.2. Absolutely conserved residues, partially residues and similar residues are shown in black, dark grey and light grey, respectively. Comparison between EgAgB8 mature peptides shows that EgAgB8/1, EgAgB8/3 and EgAgB8/5 are more closely related to each other than to EgAgB8/2 and EgAgB8/4, as shown in the alignment (B) and in the tree (C) obtained employing DrawTree from Biology Workbench 3.2. (D) Biochemical data for EgAgB8/2 and EgAgB8/3 (from sequences squared in A). The theoretical isoelectric point (pI) and the theoretical extinction coefficient were also estimated using Biology Workbench 3.2 software. Accession numbers: EgAgB8/1: AAD38373, EgAgB8/2: AAC47169, EgAgB8/3: AAK64236, EgAgB8/4: AAQ74958 and EgAgB8/5: BAE94835.