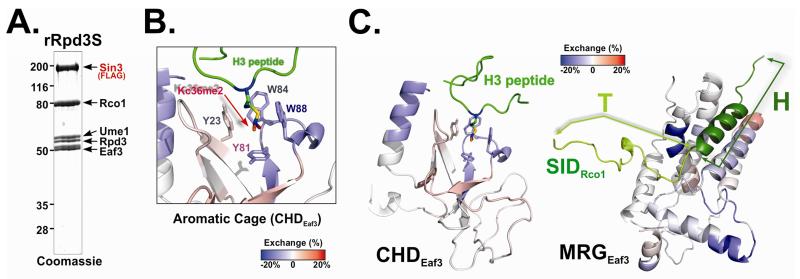
Figure 1. Rpd3S undergoes conformation changes upon Rpd3S contact with nucleosomes.
(A) Coomassie staining of the recombinant Rpd3S complex used in DXMS experiments. (B-C) Changes in the deuteration levels of Eaf3 upon Rpd3S binding to nucleosomes suggest conformational changes of Rpd3S; (B) Deuterium exchange results were mapped to 3D structure of the chromo domain of Eaf3 (PDB-2K3X). Blue color indicates slower deuterium exchange rates upon Rpd3S contact with nucleosomes, while red areas represent increased exchanges; Four aromatic residues that form the methyl-lysine binding pocket were labeled; (C) A zoom-out view of the deuterium exchange results of CHD (PDB: 2K3X) (Left) and MRG/SID (a molecular model based on PDB-2LKM using SWISS-MODEL)(the right panel). SID is represented in cartoon and green. The helix region of SID (dark green) is defined as the “H” region and the turn region (light green) is referred to as “T”. See also Figure S1.