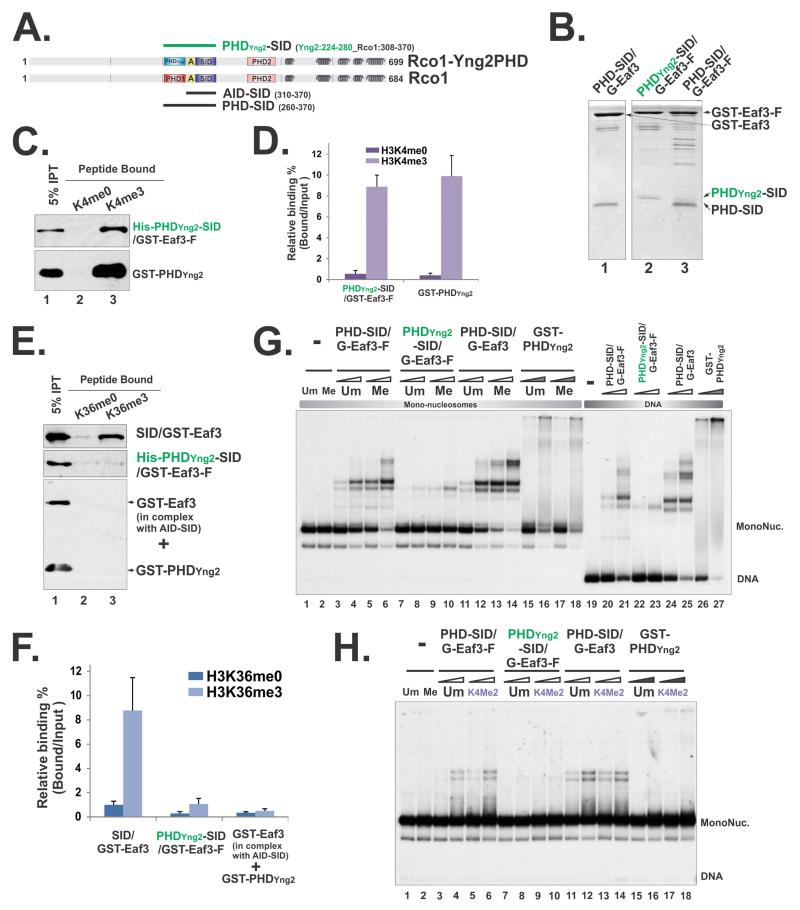
Figure 6. Perturbation of PHD domain within the minimal nucleosome binding module compromised its nucleosome engagement.
(A) A schematic illustration of the PHDYng2-SID/Eaf3 construct. “A” framed in the yellow box represents the AID domain. Amino acids that were included in the hybrid protein were indicated by the residue numbers behind each constructs. (B) Coomassie staining of tandem purified hybrid Rco1-Eaf3 heterodimers. (C) Histone peptide pull-down using unmethylated and methylated H3K4 peptides. (D) Quantification of (C) based on three repeats. Data are represented as mean ± SEM. (E) Histone peptide pull-down using unmethylated and methylated H3K36 peptides. The low level binding of PHDYng2-SID/Eaf3 was likely due to PHDYng2-somehow slightly compromises AID function, because when AID-SID/Eaf3 heterodimers and GST-PHDYng2 were mixed together, no binding was detected. (F) Quantification of (E) based on three repeats. (G) EMSA using mono-nucleosome substrates that unmethylated or tri-methylated at H3K36 and DNA. Two concentrations of heterodimers were 15pM, 30pM respectively and indicated as open triangles. 3.2μM and 6.4μM of GST-PHDYng2 were used and labeled as filled triangles. (H) EMSA using mono-nucleosome substrates that unmethylated or di-methylated at H3K4. 1.6μM and 3.2μM of GST-PHDYng2 were used.