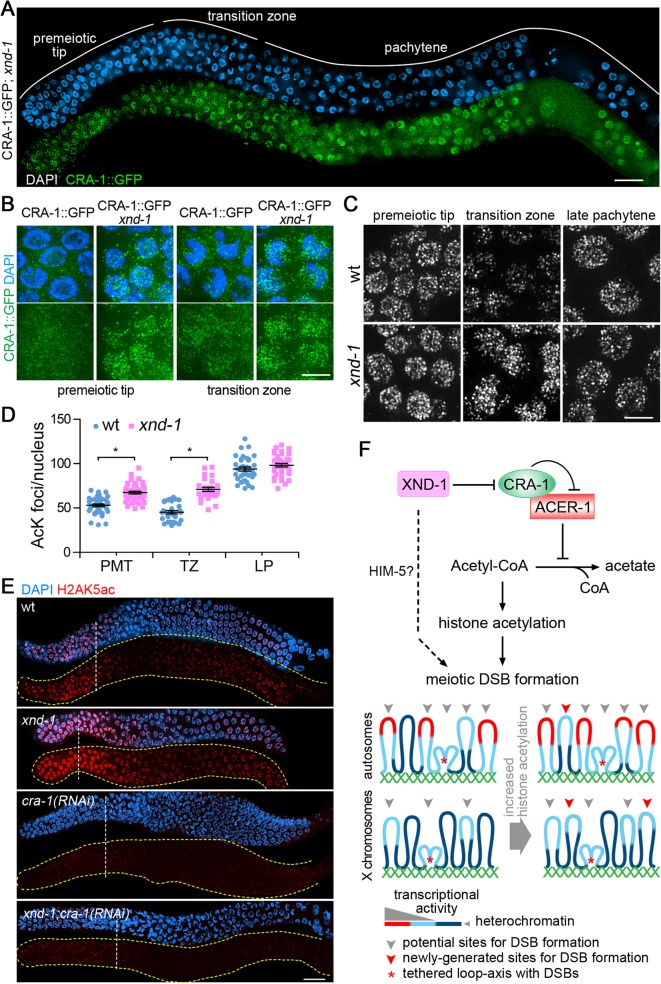
Fig 7. CRA-1 expression pattern is altered in xnd-1 mutants.
(A) CRA-1::GFP expression in xnd-1 mutant gonads. Gonads dissected from CRA-1::GFP; xnd-1 hermaphrodites were co-stained with anti-GFP antibody (green) and DAPI (blue). Bar, 20 μm. (B) High-magnification images of representative germline nuclei show the expression of CRA-1::GFP (green) at the premeiotic tip and transition zone in wild type and xnd-1 mutants. Bar, 5 μm. (C) High-magnification images of nuclei at the indicated stages from wild type and xnd-1 mutant gonads stained with an anti-acetylated lysine antibody. Images show projections through 3D data stacks of whole nuclei. Bar, 5 μm. (D) Quantification of the number of acetylation foci observed per nucleus in (D). Bars represent the mean number of foci ± SEM. * P<0.0001, by the two-tailed Mann-Whitney test, 95% C.I. (E) Gonads dissected from the indicated genotypes were co-stained with an anti-H2AK5ac antibody (red) and DAPI (blue). Bar, 20 μm. Yellow dashed lines were utilized to facilitate visualization of the gonad’s outline when only the antibody signal is depicted. White dashed vertical lines indicate exit from mitosis and entrance into meiosis. (F) A new histone acetylation-regulating protein network and sentinel chromosome model. In this regulatory network, XND-1 negatively regulates CRA-1, which interacts with and antagonizes ACER-1, an acetyl-CoA hydrolase, thereby providing a mechanism for regulating histone acetylation via modulation of acetyl-CoA levels. We propose that regulation of histone acetylation is linked to regulation of meiotic DSB formation. Dashed line: XND-1 promotes efficient meiotic DSB formation independent of its role in the histone acetylation regulatory network (potentially through HIM-5). This additional mechanism of function remains to be characterized. How are regulation of histone acetylation and meiotic DSB formation linked? Highly heterochromatic regions (dark blue) are not the preferred environments for DSB formation. Chromatin with a minimal level of histone acetylation, either transcriptionally active chromatin (red) or regions with lower histone acetylation thresholds (light blue), are better environments for DSBs. An increase in global histone acetylation does not remarkably increase the DSB-preferred chromatin regions on the autosomes, where they are already more prevalent due to their euchromatic state, but can significantly increase this type of site on the X chromosomes, where they are rare due to the highly heterochromatic environment. Our data suggests that DSBs and/or early stages of DSB repair take place close to chromosome axes. This could be either via tethering of loop sequences to axes, with DSB formation/repair taking place at axes as indicated by the red asterisk, or via breaks forming directly at sequences located near the base of the loops and in close proximity to axes, without any tethering (not shown). While we cannot exclude the latter, we favor the former given the evidence of chromatin silencing following DSB formation ([38]; Fig. 5B).