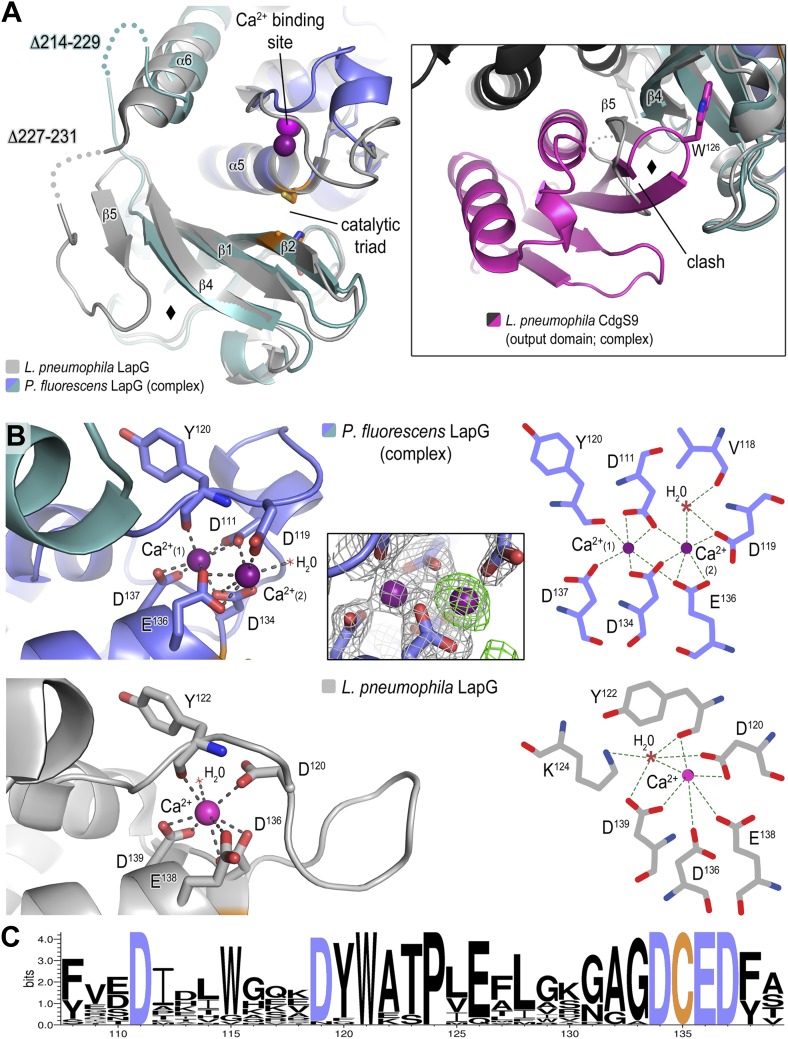
Figure 5. Structure of P. fluorescens LapG in complex with CdgS9Output.
(A) The structure of L. pneumophila LapG shown in gray (PDB: 4FGO, Chatterjee et al., 2012) was superimposed on that of P. fluorescens LapG, colored in slate and cyan, when in complex with CdgS9Output. A close-up view of the differences observed in the C-terminal lobe of the two LapG structures is shown (right inset). Disordered regions not resolved in the crystal structures are shown as dots. Calcium ions are represented as magenta spheres. The catalytic triad residues (C-H-D) are shown as sticks with the carbon atoms colored in orange. (B) The Ca2+ binding sites of LapG. P. fluorescens (top panel, blue cartoon) and L. pneumophila LapG (bottom panel, gray cartoon) coordinate two or one calcium ion, respectively. In P. fluorescens LapG, an Fo–Fc omit map contoured at 3σ (green mesh, inset of top panel) shows strong evidence for a second calcium ion not seen in the L. pneumophila structure, which is made possible by local conformational differences in the so-called calcium binding loop that bring D111 of P. fluorescens into position to coordinate the additional calcium ion. A LIGPLOT (Wallace et al., 1995) (right panels) depicts the coordination of calcium ions. (C) Conservation of the di-calcium binding site. A WebLOGO plot (Crooks et al., 2004) generated from an alignment of 18 LapG ortholog sequences demonstrates the conservation of coordinating residues (blue font) as well as the nucleophilic cysteine (yellow font).