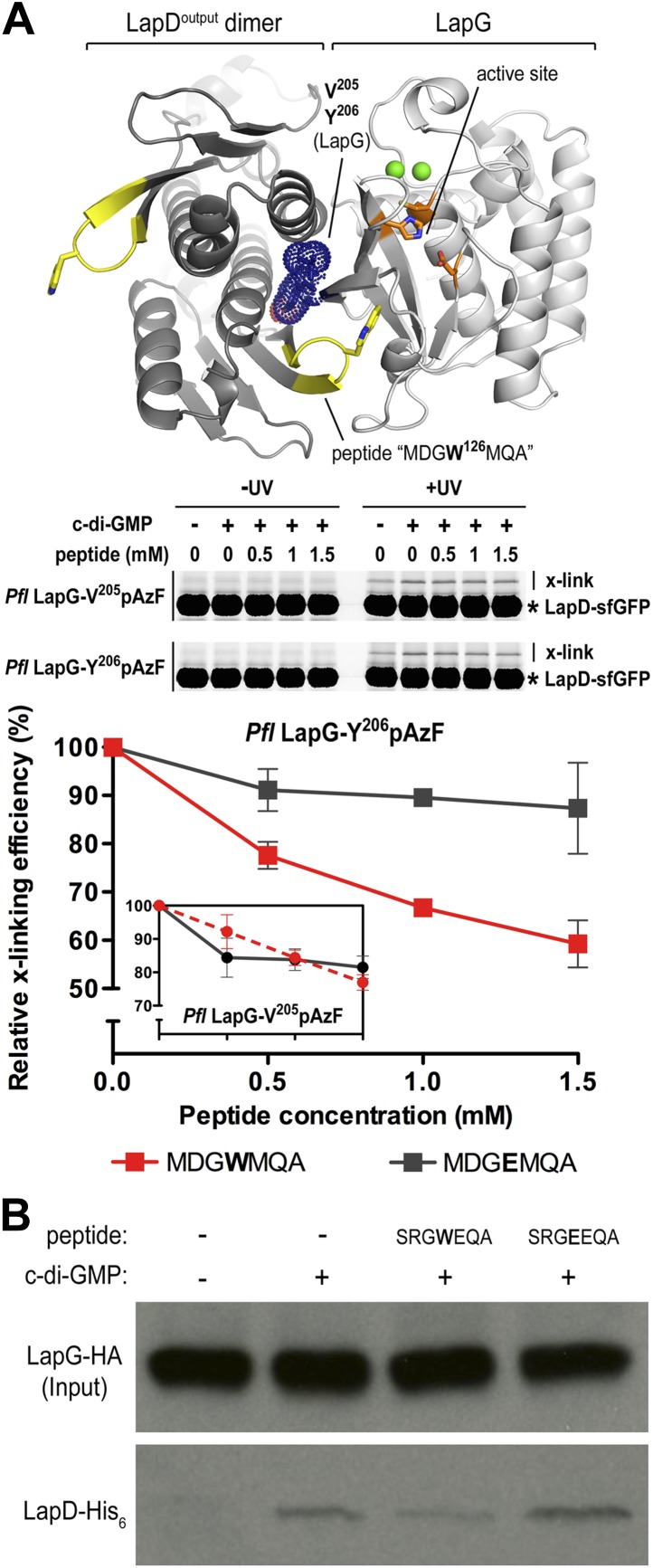
Figure 8. Attenuating LapD–LapG interactions with a peptide containing the GWxQ binding motif.
(A) Cross-linking assay using purified proteins. The CdgS9Output-LapG crystal structure depicts the GWxQ β-hairpin motif of CdgS9Output (yellow cartoon) as well as the two sites of pAzF incorporation (blue mesh) on LapG used to assess LapD–LapG interactions (top panel). To assess the effects of the MDGW126MQA peptide on LapD–LapG interactions, detergent-solubilized, full-length LapD-sfGFP was incubated with purified non-fluorescent LapG containing pAzF at V205 (inset in bottom panel) or Y206 (main graph in bottom panel), and increasing concentrations of peptide. Half of the sample was subjected to UV irradiation for 5 min prior to analysis by SDS-PAGE. The average intensity of the fluorescent band corresponding to LapD-sfGFP crosslinked to LapG from three independent measurements was normalized to non-crosslinked LapD-sfGFP, averaged, and plotted as a function of peptide concentration (±SD) (red line; bottom panel). The peptide MDGE126MQA was used as a negative control (gray line; bottom panel). (B) Pull-downs of LapG-HA from lysates of P. fluorescens. LapD-His6 and LapG-HA were co-expressed in a P. fluorescens strain with deleted lapD and lapG genes. Western blots using HA (top panel) and His6 (bottom panel) specific primary antibodies were used to assess the levels of LapD-His6 that co-purify with LapG-HA in the presence or absence of peptides and c-di-GMP.