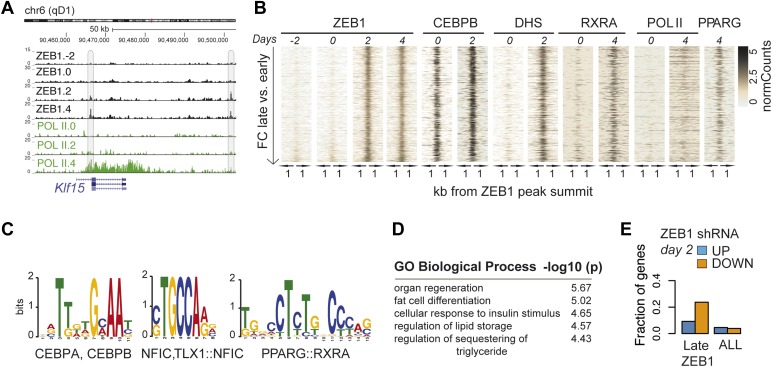
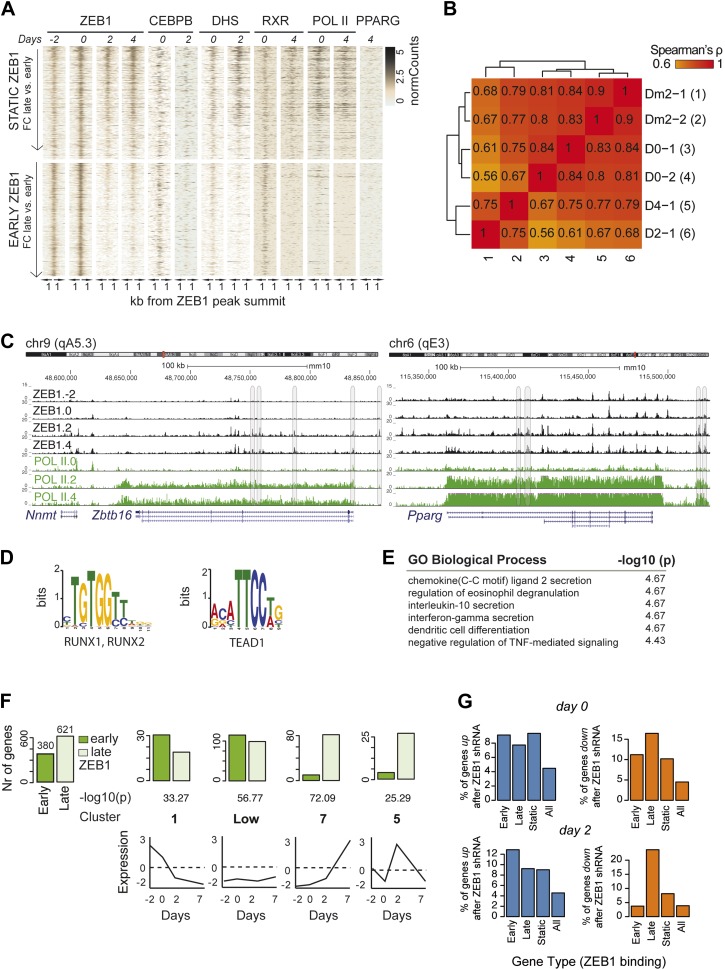
Figure 4. ZEB1 binding increases at adipogenic genes during 3T3-L1 differentiation.
(A) ZEB1 and POLII read density tracks at the Klf15 locus during 3T3-L1 differentiation (days −2, 0, 2, and 4). Late-only bound regions are highlighted. (B) ZEB1, C/EBPβ, RXRα, PPARγ, POLII normalized ChIP (‘Materials and methods’) as well as DHS enrichments in a 2 kb window around the summits of late-only (days 2 and 4 but not days −2 and 0; padj ≤ 0.1, FC ≥ 2) ZEB1-bound regions during 3T3-L1 differentiation. (C) Differential motif discovery using MEME and a 50 bp sequence centered on summits of late-only vs. static ZEB1 peaks reveals adipogenic motifs: C/EBPα|C/EBPβ, NFIC and PPARG::RXR (p < 10−3, ‘Materials and methods’). (D) GREAT-based (McLean et al., 2010) Gene Ontology enrichment analysis of genes associated with late-only vs. static ZEB1 binding reveals terms associated with fat cell differentiation and function (complete results in Supplementary file 1G [Gubelmann et al., 2014]). (E) Fraction of genes associated with late-only ZEB1 binding and fraction of all genes significantly up (blue) and down (orange)-regulated after ZEB1 KD as measured at differentiation day 2 (complete results in Figure 4—figure supplement 1G).