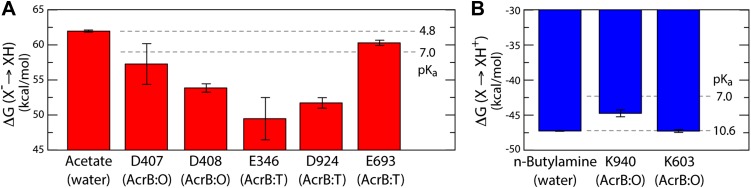
Figure 6. Free energy of protonation for selected glutamate, aspartate and lysine residues in the L, T and O states.
All values were computed via all-atom molecular simulations in a phospholipid membrane (‘Materials and methods’). Computed values for side-chain analogs in solution, whose pKa is known experimentally, provide a means to translate the computed free-energy values into a pKa scale (dashed lines). (A) The calculations indicate upward shifts in the pKa of E346 and D924 in the T state, and of D407 and D408 in the O state, relative to acetate in water, whose magnitude indicate that in those conformational states these residues are protonated at physiological pH. By contrast, the pKa of E693 is hardly shifted, consistent with its location on a water-exposed, electrostatically neutral area of the protein surface. (B) The pKa of K940 in the O state is also shifted downwards relative to n-butylamine in solution, on account of its being largely unexposed, but does not become deprotonated owing to polar contacts with N941 and T978 (Figure 4).