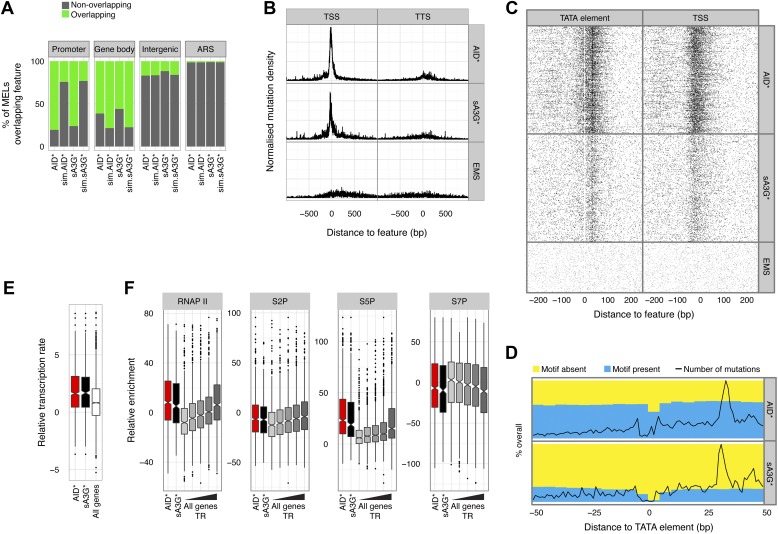
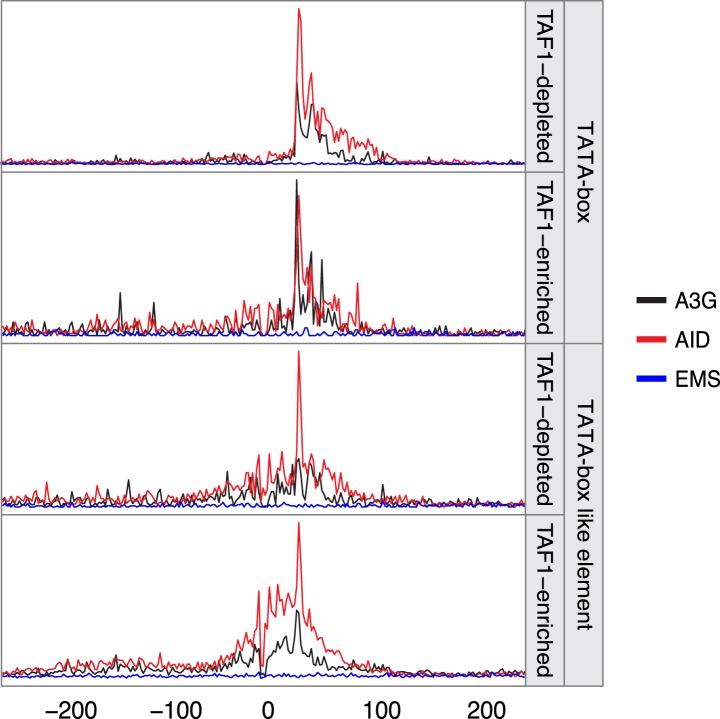
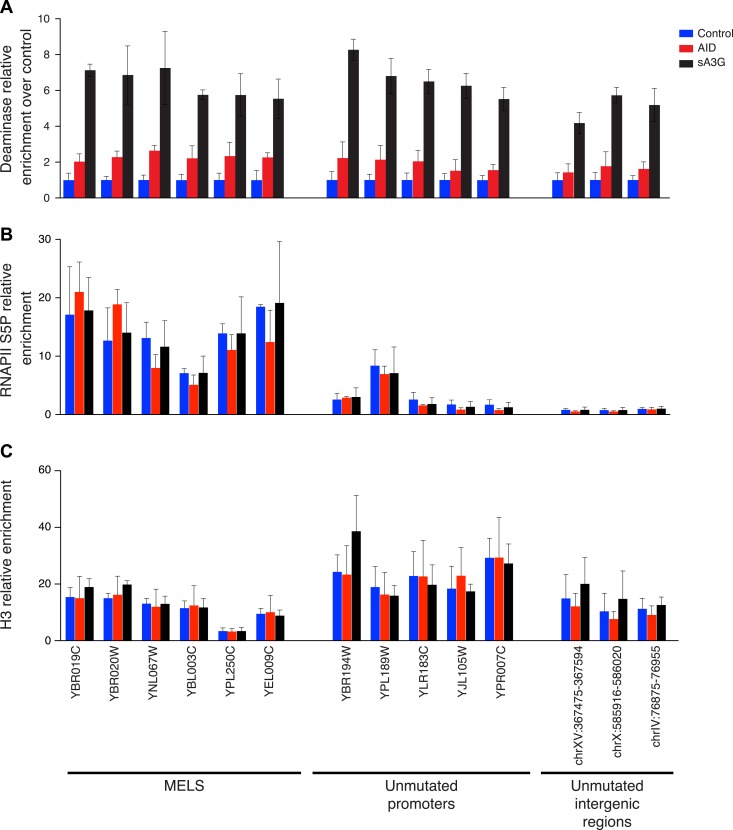
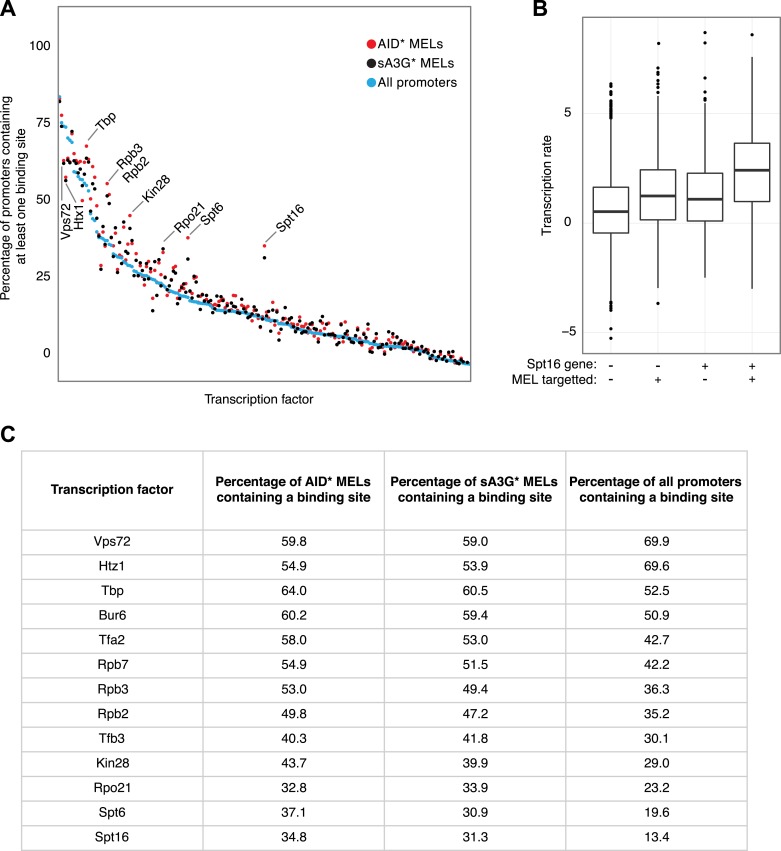
Figure 3. Deaminase mutation footprints are focussed to the pre-initiation complex region of active promoters.
(A) Proportion of promoters, gene bodies, intergenic regions and replication origins (ARS) harbouring a MEL (green) or not (grey) for AID* and sA3G* datasets vs the expected distribution (sim.AID*sA3G*) determined by Monte Carlo simulation of equivalent sized fragments for each MEL dataset distributed randomly across the genome. (B) Density of mutations in relation to their distance to the nearest transcription start site (TSS) of mRNA (RNAP II) transcripts compared to the density relative to transcription termination sites (TTS). Data includes all mutations in addition to MELs. (C) Deaminase mutations relative to the TATA or TATA-like element for each RNAP II promoters (Rhee and Pugh, 2012) compared to the mutation distance distribution aligned to the transcription start site (TSS). (D) Proportion of AID* or sA3G* mutable motifs within RNAP II promoter regions, centred on the TATA-elements (Rhee and Pugh, 2012). Total number of mutations for each dataset is shown at each position (black line). (E) Relative transcription rates (see methods) at RNAP II promoters targeted by MELs compared to relative transcription rates for all RNAP II genes in gal induced conditions (García-Martínez et al., 2004). (F) Relative enrichment of RNAP II and RNAP II CTD phosphorylation (S2P, S5P and S7P) in promoters containing AID* (red) and sA3G* (black) MELs and all RNAP II promoters (grey) ranked according to transcriptional activity (García-Martínez et al., 2004).