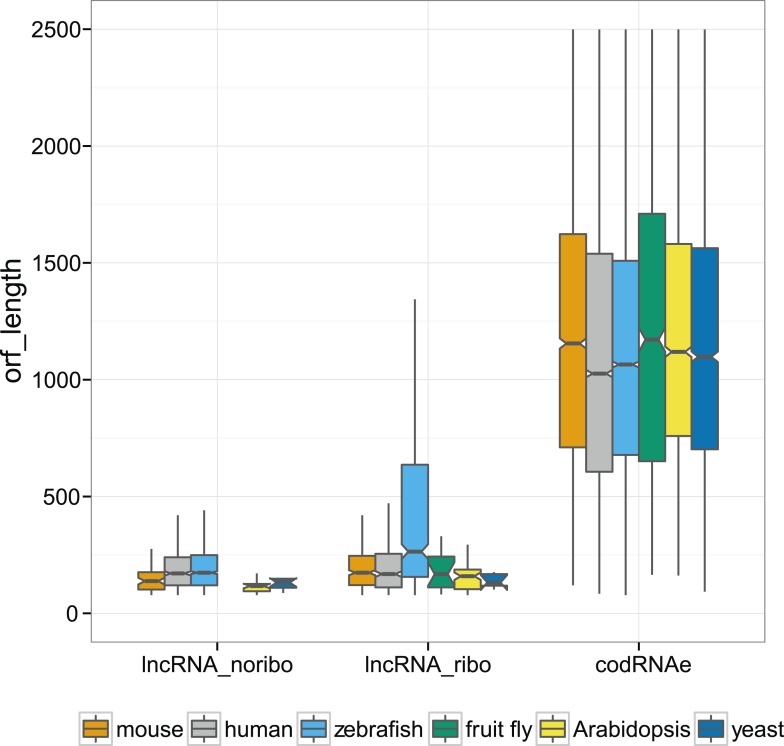
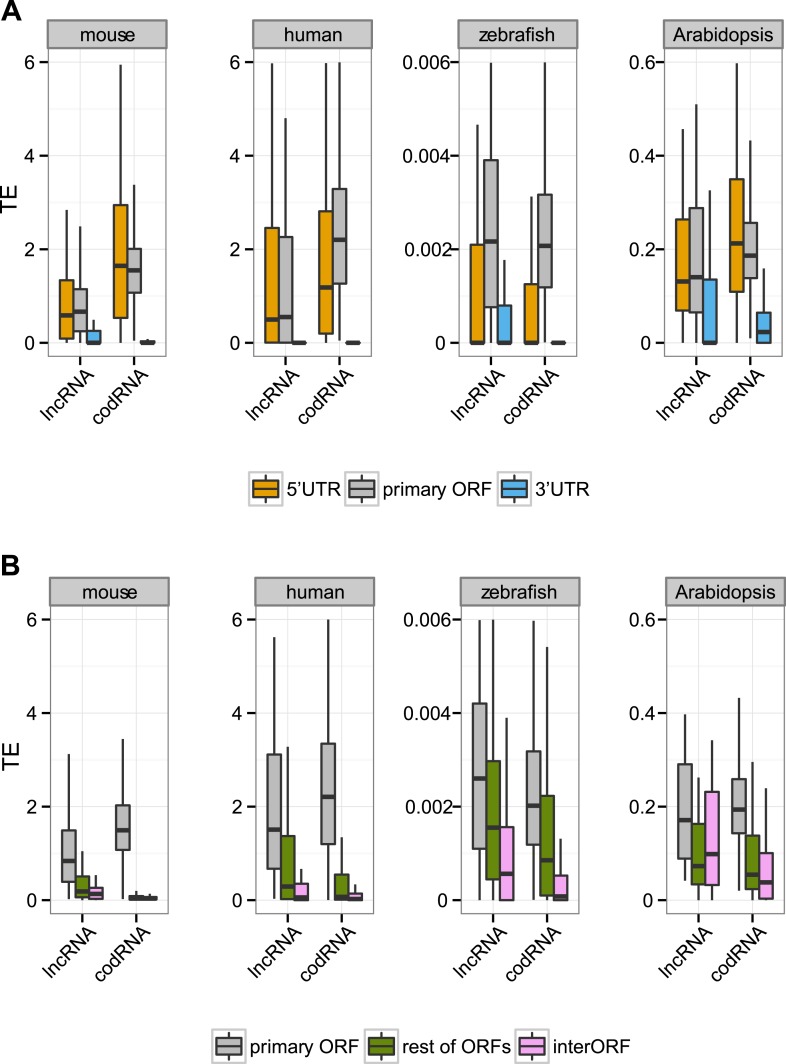
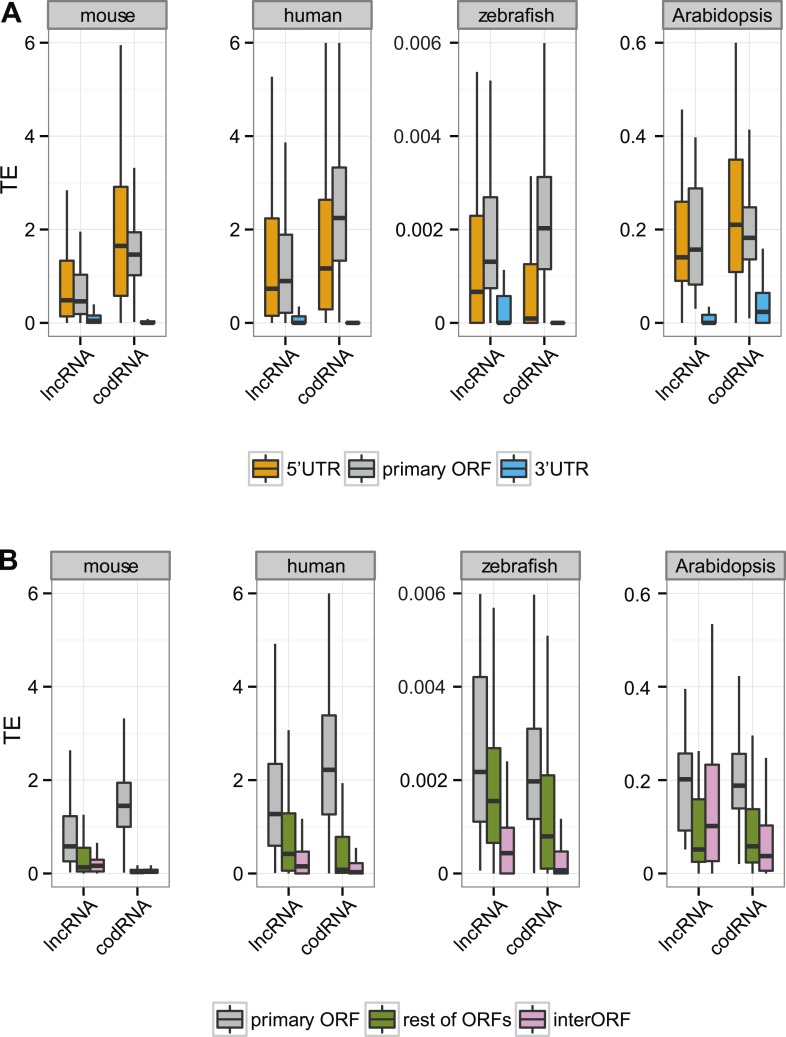
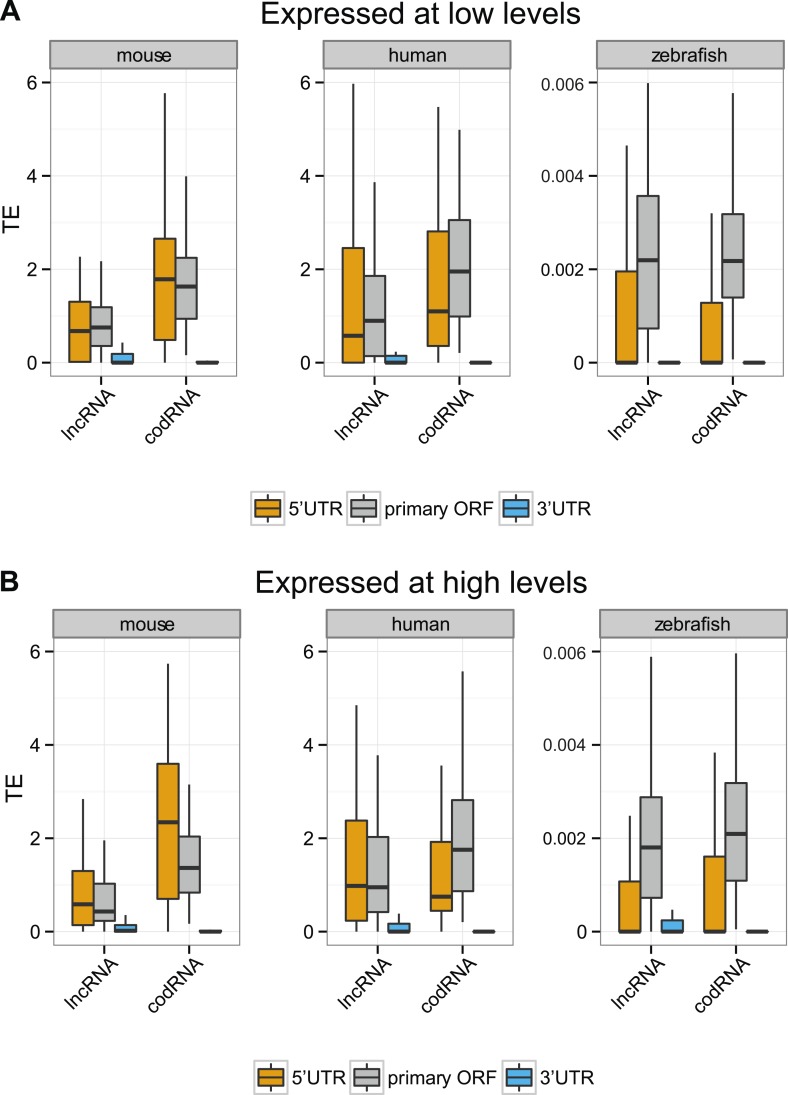
Figure 5. Ribosome association in different transcript regions.
(A) Density plot of the relative length of the primary ORF in lncRNA_ribo and codRNA with respect to transcript length. For comparison data for the longest ORF in lncRNA_noribo is also shown (except for fruit fly due to insufficient data). (B) Box-plots of TE distribution in primary ORF, 5′UTR, and 3′UTR regions. The area within the box-plot comprises 50% of the data, and the line represents the median value. The analysis considered all transcripts with 5′UTR and 3′UTR longer than 30 nucleotides and >0.2 FPKM in all three regions. The number of transcripts was 1956 codRNA and 159 lncRNA_ribo in mouse, 3558 codRNA and 139 lncRNA_ribo in human, 5216 codRNA and 252 lncRNA_ribo in zebrafish, and 2019 codRNA and 33 lncRNA_ribo in Arabidopsis. (C) Box-plots of TE distribution in primary ORFs, rest of ORFs with ribosome profiling reads and non-ORF regions (interORF). The analysis considered all transcripts with at least two ORFs and more than 30 nucleotides interORF. The number of transcripts was 3264 codRNA and 204 lncRNA_ribo in mouse, 3104 codRNA and 168 lncRNA_ribo in human, 1646 codRNA and 212 lncRNA_ribo in zebrafish, and 1098 codRNA and 25 lncRNA_ribo in Arabidopsis. Fruit fly and yeast were not included in the last two analyses due to insufficient data (<8 lncRNA_ribo meeting the conditions).