Figure 1.
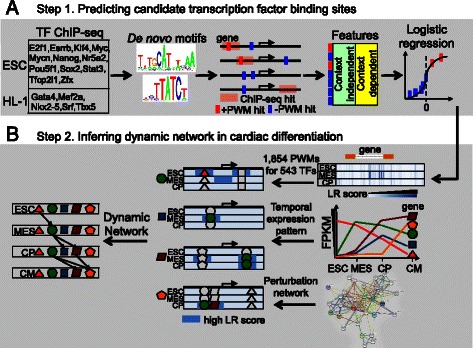
Overview of our two-step strategy to infer dynamic regulatory networks during cardiac differentiation. (A) Training a general logistic regression model to predict the probability being bound by any transcription factor (LR score) in 40 kb cis-regions surrounding transcriptional start sites (TSS) of expressed genes. The response variables of the model indicate whether the hit of the PWM of a specific TF coincides with the peak region of the corresponding ChIP-seq data. 15 context-independent (e.g. conservation) and 54 context-dependent (e.g. mean intensity of H3K27ac in 1 kb surrounding any base) features were used to train the logistic regression model. (B) Context-dependent LR score, temporal expression profiles and perturbation networks were used to infer the dynamic regulatory networks based on a time-varying dynamic Bayesian network model.
