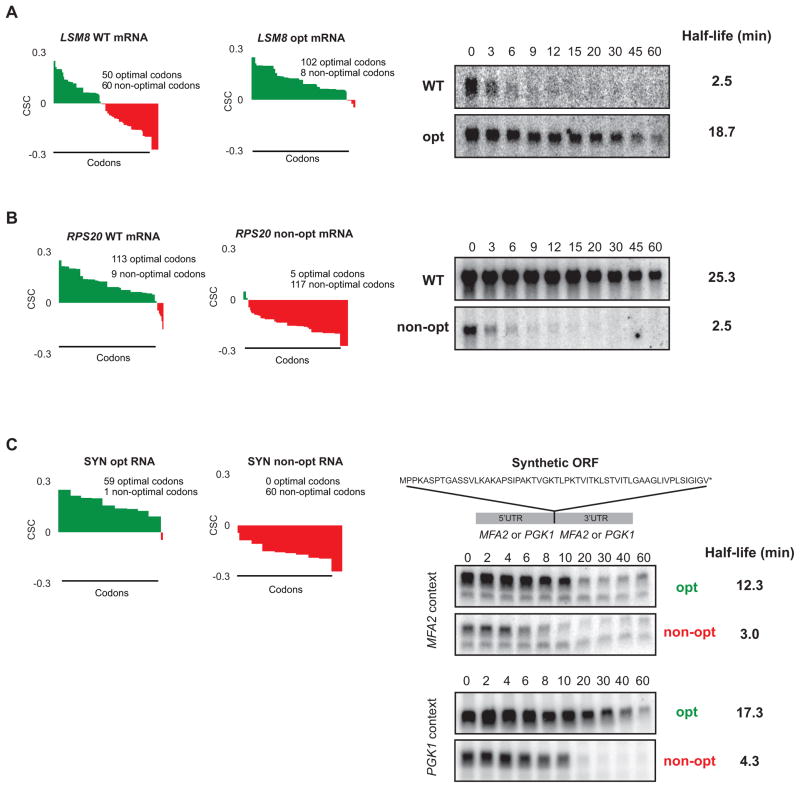
Figure 4. Stability of mRNAs can be controlled by altering codon optimality.
(A) Codon optimality diagram of LSM8 (as Fig. 3E), a naturally non-optimal mRNA shown. LSM8 OPT is a synonymously substituted version of LSM8 engineered for higher optimality. Northern blots of rpb1-1 shut-off experiments are shown on the right with half-life of both reporters. Quantitation is normalized to SCR1 loading controls not shown. (B) As (A), except a naturally optimal mRNA, RPS20 (as in Fig. 3D), has been engineered for lower optimality as RPS20 non-opt. Northern blots of rpb1-1 shut-off experiments are shown on the right with half-life of both messages. Quantitation is normalized to SCR1 loading controls not shown. (C) Codon optimality diagrams showing a synthetic mRNA (SYN) encoding the polypeptide shown. Peptide is artificially engineered and has no similarity to any known proteins. SYN opt and non-opt were both inserted into flanking regions from a stable transcript (PGK1) and unstable transcript (MFA2). Northern blots on the right show GAL shut-off experiments demonstrating stability of the SYN mRNA in context of the MFA2 and PGK1 flanking sequences. Quantitation is normalized to SCR1 loading controls not shown. See also Figure S3.