Figure 1.
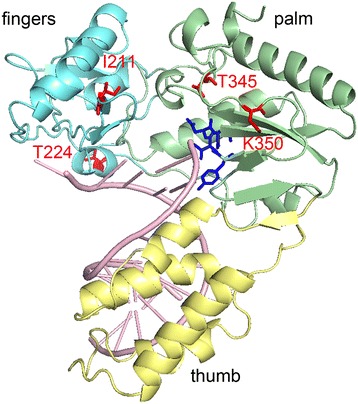
Localization of the AZT resistance substitutions. The putative locations of the four substituted amino acid residues in the highly resistant mt4 (I211, T224, T345, K350) are highlighted in red in a structural model of the SFVmac RT polymerase domain. The polymerase active site motif YVDD is highlighted in blue. The model is based on the structure of XMRV RT (PDB: 4HKQ) with an RNA/DNA hybrid substrate and was generated using the Program SWISS-MODEL [25].
