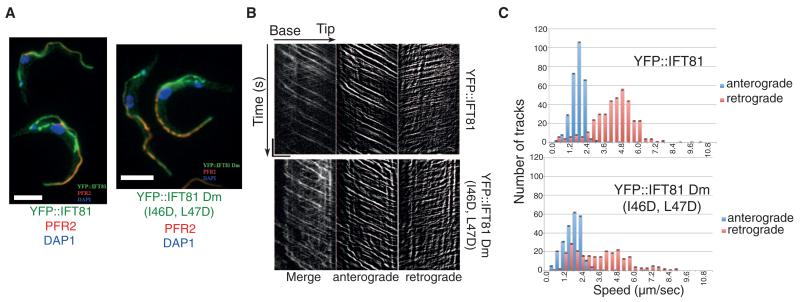
Fig. 3. IFT81N is not required for normal IFT.
(A) Immunofluorescence analysis of methanol-fixed trypanosomes expressing the indicated YFP fusion proteins from the endogenous locus stained with an antibody to green fluorescent protein (GFP) (green) and with the antibody to PFR2 L8C4 to visualize the flagellum (red). The left panel corresponds to a control strain expressing YFP::IFT81 and the right panel to the mutant YFP::IFT81Dm, where the IFT81N CH domain is unfolded. Scale bar, 5 μm. (B) Kymograph generation and separation of anterograde and retrograde traces. Kymographs were extracted from videos of cells expressing YFP::IFT81 (movie S1) or YFP::IFT81Dm (movie S2). Panels show the complete kymograph, anterograde events, and retrograde events (from left to right). The x axis corresponds to the length of the flagellum (horizontal scale bar, 5 μm) and the y axis to the elapsed time (vertical scale bar, 5 s). (C) Quantitation of the kymograph analysis shown in (B). Anterograde (blue) and retrograde velocity (red) distribution of IFT particles are calculated from cells expressing YFP::IFT81 and YFP::IFT81Dm. The kymographic analysis reveals robust anterograde trafficking with a speed of 1.75 ± 0.55 μm/s for YFP::IFT81 (n = 294 tracks from 15 cells) and 1.68 ± 0.72 μm s−1 for YFP::IFT81Dm (n = 244 tracks from 15 cells). These values are in line with those reported for anterograde movement of GFP-IFT52 (15). Curiously, retrograde transport was slowed down in the case of YFP::IFT81Dm, where a second population of relatively slow trains was detected.