Fig. 3.
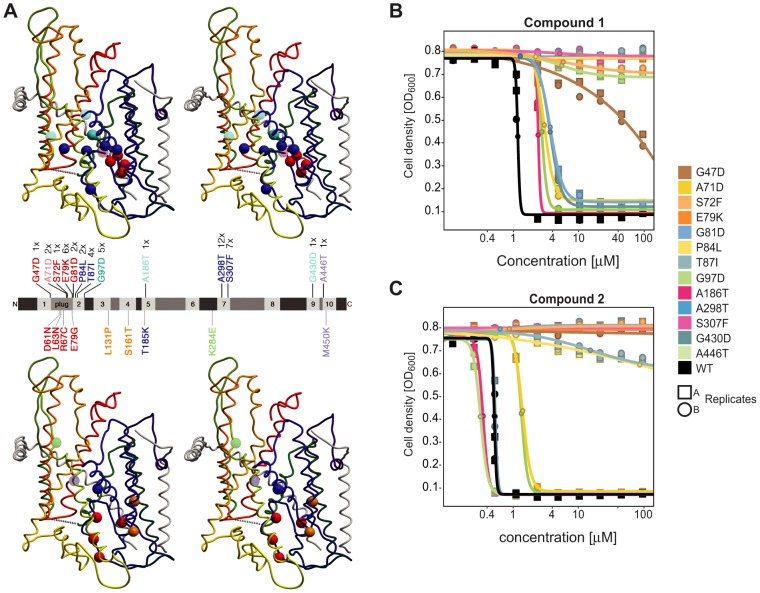
Selection of yeast mutants resistant to the inhibitors. (A) Thirteen different single amino acid mutations were identified in SEC61 of chemically mutagenized drug-efflux-compromised yeast cells selected for resistance to 30 µM compound 1 on plates. Their positions are indicated in the model of the yeast Sec61 complex (Junne et al., 2006) shown in stereo in the top panel. Below, previously studied Sec61p single point mutants that were found to be resistant (see Table 2) are similarly presented. In the middle, the Sec61p sequence is schematically shown as a bar with numbered transmembrane domains. The newly selected and the old resistance mutations are indicated above (with their frequency of occurrence) and below, respectively. Mutations resistant to both compounds 1 and 2 are shown in red if they have a prl phenotype, and in blue if not. Mutations resistant only to compound 1 (not prl) are shown in cyan, those resistant only to compound 2 in green (not prl) or orange (prl). The weak mutants are indicated by lighter shades. (B,C) Dose–response curves of wild-type yeast strains in liquid cultures expressing the indicated Sec61p point mutants for compound 1 and compound 2, respectively. Moderate to high resistance to compound 1 (against which they were originally selected) was validated for all mutations. They were also resistant to compound 2, with the notable exception of G97D, A186T and G430D. Resulting IC50 and r2 values are listed in Table 1. Boxes and circles indicate independent replicate experiments. Dose–response curves for resistant mutants shown in the lower part of A are presented in supplementary material Fig. S3.