The majority of chronic lymphocytic leukemia (CLL) patients are diagnosed with early-stage disease but the currently used prognostic tools appear to be less informative in this group of patients.1 This is especially problematic for patients with mutated immunoglobulin genes (M-CLL) as they have a more diverse clinical course when compared with patients with unmutated immunoglobulin genes (U-CLL).1, 2, 3, 4 Given the emergence of promising targeted, less toxic, therapeutics in CLL,5, 6 there is an increased need to identify patients who might benefit from early treatment with these new agents.
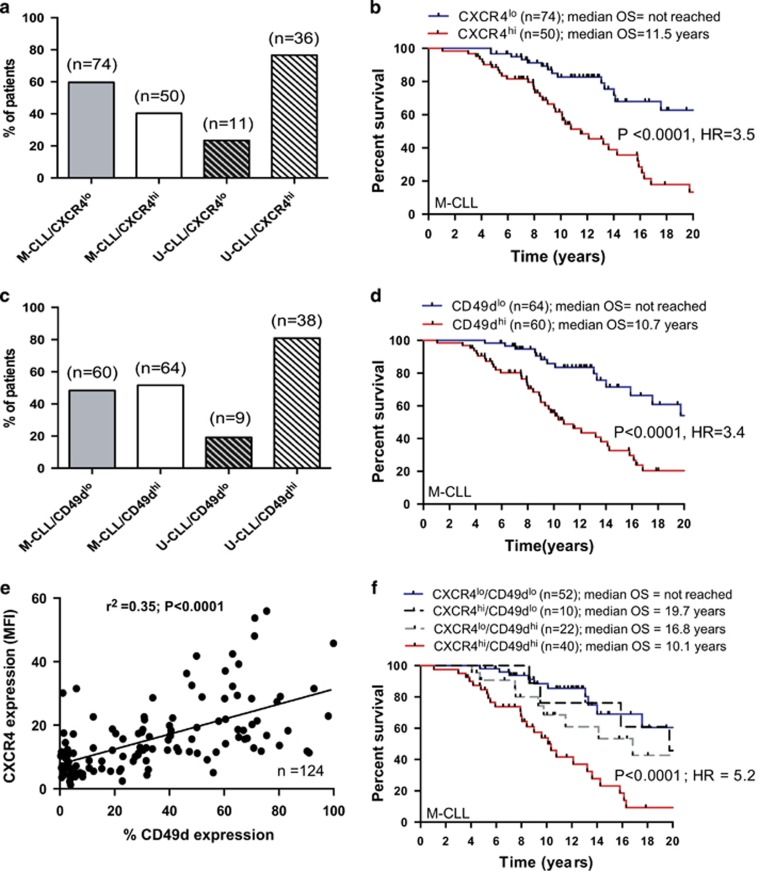
Chemokine receptors expressed on CLL cells are thought to regulate the trafficking of the leukemic cells between blood and lymphoid tissues.5 Logically, the tendency of CLL cells to return to tissue sites where they are cytoprotected and are driven to proliferate contributes to clinical aggressiveness. It is therefore possible that these receptors represent promising prognostic markers and potentially useful therapeutic targets. We previously reported that one such chemokine receptor, CXCR4, is associated with poor clinical outcome in an unselected cohort of CLL patients.7 Here we specifically focused our attention on M-CLL samples to ascertain the impact of CXCR4 expression in this clinically heterogeneous subset. The cohort was made up of 60 M-CLL patients from the original study and 64 additional M-CLL patients. The patient characteristics are given in Supplementary Table 1. CXCR4 expression was determined using the three-color flow cytometric assay described previously.7 We used the median expression as a binary classifier based on receiver operating characteristic analysis. Of the 124 M-CLL samples analyzed, 50 (40.4%) were classified as CXCR4hi and 74 (59.6%) as CXCR4lo; M-CLL patients showed much more heterogeneity in CXCR4 expression than U-CLL (Figure 1a). Importantly, elevated CXCR4 expression in M-CLL was a strong determinant of reduced overall survival (Figure 1b; hazard ratio (HR)=3.5). As M-CLL patients utilizing IGHV3-21 genes have been shown to have an inferior clinical outcome,8, 9 we asked whether the CXCR4 expression was significantly different in the IGHV3-21 subset. We found no significant increase in CXCR4 expression in IGHV3-21 samples when compared with samples utilizing other IGHV gene segments (Supplementary Figure 1; P=0.92). Similarly, we found no association between CXCR4 expression and the high-risk cytogenetic risk groups (Supplementary Figure 1; P=0.41).
Figure 1.
Differential expression and prognostic impact of CXCR4 and CD49d in M-CLL. (a) We analyzed the expression of CXCR4 in a cohort of 124 serially collected M-CLL samples and used the median expression as a binary classifier based on receiver operating characteristic analysis. M-CLL patients showed a much more heterogeneous expression of CXCR4 than U-CLL cases. (b) M-CLL patients with a CXCR4hi phenotype had a significantly inferior outcome when compared with CXCR4lo patients. (c) The cohort was then categorized into CD49dlo (<30% expression) and CD49dhi (⩾30% expression) subsets. Again M-CLL cases showed much more heterogeneity in the expression of CD49d than U-CLL cases. (d) M-CLL patients with a CD49dhi phenotype had a significantly inferior outcome when compared with CD49dlo patients. (e) When assessed as continuous variables, CXCR4 and CD49d were strongly correlated. Similarly, using categorical cutoffs to define the cohort, the majority of M-CLL cases showed concordant expression for CXCR4 and CD49d: CXCR4hi/CD49dhi or CXCR4lo/CD49dlo. 27% of the cases were discordant for these markers. (f) The Kaplan–Meier curves for the discordant cases bisected the CXCR4hi/CD49dhi and CXCR4lo/CD49dlo curves highlighting the prognostic importance of both CXCR4 and CD49d and suggesting that the combined assessment of CXCR4 and CD49d has clinical utility.
We recently showed a strong association between CXCR4 expression and CD49d in an unselected cohort of U-CLL and M-CLL patients.7 We therefore examined the expression of CD49d in this M-CLL subset using the same flow cytometric methodology and determined its prognostic relevance. In concordance with our CXCR4 data, there was heterogeneous expression of CD49d with 60 (47.4%) CD49dhi and 64(52.6%) CD49dlo. Again this heterogeneity was in marked contrast to U-CLL cases (Figure 1c). Furthermore, CD49dhi patients (>30% positive) had a significantly worse clinical outcome than CD49dlo patients (Figure 1d; HR=3.4) reinforcing the credentials of CD49d as a prognostic marker in the M-CLL subset.10 When assessed as continuous variables, CXCR4 and CD49d were strongly correlated (Figure 1e; P<0.0001). Using categorical cutoffs to define the cohort, the majority of M-CLL cases showed concordant expression for CXCR4 and CD49d: CXCR4hi/CD49dhi or CXCR4lo/CD49dlo. However, 27% of the subset was discordant for these markers (Supplementary Figure 2). The Kaplan–Meier curve for the discordant cases bisected the CXCR4hi/CD49dhi and CXCR4lo/CD49dlo curves highlighting the prognostic importance of both CXCR4 and CD49d and suggesting that the combined assessment of CXCR4 and CD49d has clinical utility. This was supported by the observation that the combination of CXCR4 and CD49d was a more powerful prognostic tool than either marker alone for the concordant cases (Figure 1f; HR=5.2). It is worthy of note that CXCR4hi/CD49dhi M-CLL cases had a similar clinical outcome when compared with U-CLL cases (Supplementary Figure 3).
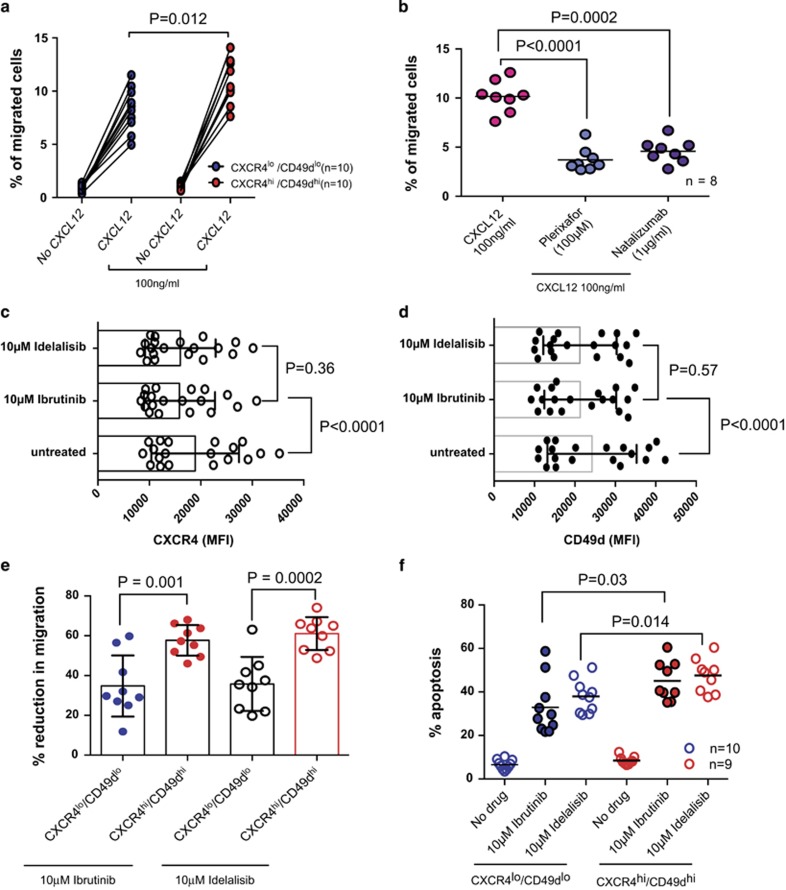
We went on to establish the functional significance of CXCR4 and CD49d expression in M-CLL. CXCR4hi/CD49dhi samples showed significantly increased migration in response to CXCL12 in transwell experiments (Figure 2a; P=0.012), which could be inhibited by both plerixafor (CXCR4 antagonist) and natalizumab (anti-CD49d antibody) even in the presence of CXCL12 (Figure 2b). We observed a significant reduction in CXCR4 expression when CLL cells were incubated with CXCL12 presumably owing to the internalization of the receptor (Supplementary Figure 4a). No change in CD49d expression was seen under these conditions (Supplementary Figure 4b). There is now a growing appreciation that the propensity of CLL cells to home to lymphoid tissues contributes to the clinical course of CLL.11, 12 We recently showed that CXCR4hi and CD49dhi samples have a higher propensity to undergo transendothelial migration in a dynamic in vitro model of the peripheral vasculature of CLL suggesting that these molecules contribute to this process.12 The importance of CLL cell migration is perhaps best exemplified by the remarkable clinical effect of the BTK and PI3Kδ inhibitors ibrutinib and idelalisib.13 Both drugs promote a redistribution of CLL cells into the peripheral vasculature from lymphoid tissues characterized by a rapid increase in peripheral lymphocytosis. These cells appear then to be trapped in the peripheral vasculature where, in the absence of prosurvival and proproliferative signals, they are potentially more susceptible to spontaneous apoptosis and chemotherapeutic agents. Although these drugs are currently being trialed in patients with progressive or advanced stage disease,14, 15 their side-effect profiles are modest and so it seems likely that they will be considered as frontline treatment options in early-stage asymptomatic patients. This being the case, it will be crucial to accurately identify those patients who might benefit from early treatment and equally those who may not require treatment at all. Intriguingly, a recent study indicated that M-CLL patients had an inferior response to ibrutinib when compared with U-CLL patients.6 Here we show that both drugs caused a significant reduction in CXCR4 expression (Figure 2c) and CD49d expression (Figure 2d). Furthermore, despite the enhanced migratory properties of CXCR4hi/CD49dhi M-CLL samples ibrutinib and idelalisib induced a significantly greater percentage reduction in the migration of CXCR4hi/CD49dhi M-CLL samples (mean migration 9.44±2.08% to 4.00±1.12%) when compared with CXCR4lo/CD49dlo samples (mean migration 8.75±1.61% to 4.97±0.85% Figure 2e). In addition, CXCR4hi/CD49dhi M-CLL samples were more susceptible to the apoptotic effects of ibrutinib and idelalisib (Figure 2f). These findings suggest that CXCR4hi/CD49dhi M-CLL patients are more dependent on BTK and PI3Kδ signaling, and hence could particularly benefit from treatment with inhibitors of these kinases.
Figure 2.
CXCR4hi/CD49dhi M-CLL cases are functionally distinct from CXCR4lo/CD49dlo M-CLL cases. In vitro migration assays were performed using 3.0-μm pore size transwell migration plates (Costar, Corning, NY, USA). A total of 106 CLL cells in 500 μl of RPMI containing 10% fetal calf serum (FCS) were added to the upper chamber of the transwell insert. 100 ng/ml of CXCL12 was added to the lower wells. The plates were incubated for 4 h at 37 °C in 5% CO2. Controls without CXCL12 were maintained in each experiment to account for passive diffusion of cells (<1.5% in every case). (a) CXCR4hi/CD49dhi samples (n=10) showed significantly increased migration in response to CXCL12 when compared with CXCR4lo/CD49dlo samples (n=10). (b) Addition of plerixafor (100 μM) or natalizumab (1 μg/ml) to the upper chamber of the transwell insert significantly inhibited CLL cell migration despite the presence of a CXCL12 chemokine gradient (n=8). Exposure to ibrutinib or idelalisib for 24 h resulted in a significant reduction in (c) CXCR4 expression and (d) Cd49d expression in the viable CD19+/CD19+ CLL cells. (e) Although CXCR4 hi/CD49dhi samples were inherently more migratory than CXCR4lo/CD49dlo samples, incubation with ibrutinib (10 μM) and idelalisib (10 μM) for 4 h induced a significantly larger percentage reduction in migration in CXCR4hi/CD49dhi samples. (f) CLL cells derived from M-CLL patients were maintained in vitro in RPMI supplemented with 10% FCS and 5 ng/ml interleukin-4 in the presence or absence of 10 μM ibrutinib. After 48 h apoptosis was assessed using Annexin V and propidium iodide labeling. CXCR4hi/CD49dhi samples (n=9) showed significantly increased apoptotic cell death in response to ibrutinib and idelalsib when compared with CXCR4lo/CD49dlo samples (n=10). All paired and unpaired observations were analyzed using the Student's t-test after confirming that the data were Gaussian or a Gaussian approximation using the omnibus K2 test.
Taken together, our data provide evidence that CXCR4 and CD49d are important modulators of prognosis in M-CLL patients. Furthermore, the combination of CXCR4 and CD49d measurement provided even greater prognostic resolution for concordant M-CLL cases. This knowledge could facilitate enhanced flow cytometry-based risk stratification for this heterogeneous group of patients and identify those who might particularly benefit from BTK or PI3Kδ inhibitors or therapeutics designed to target the function of CXCR4 and CD49d. Similarly, it could delineate a substantial population of CXCR4lo/CD49dlo M-CLL patients with very low risk of disease progression who could be monitored less frequently.
Acknowledgments
This work was supported in part by grants from Leukaemia & Lymphoma Research and the Leukaemia Research Appeal for Wales. CP is also supported by the National Institute for Social Care and Health Research (NISCHR) through the Cancer Genetics Biomedical Research Unit and FS is supported by the CLL Global Research Foundation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
Supplementary Material
References
- Pepper C, Majid A, Lin TT, Hewamana S, Pratt G, Walewska R, et al. Defining the prognosis of early stage chronic lymphocytic leukaemia patients. Br J Haematol. 2012;156:499–507. doi: 10.1111/j.1365-2141.2011.08974.x. [DOI] [PubMed] [Google Scholar]
- Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–1854. [PubMed] [Google Scholar]
- Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–1847. [PubMed] [Google Scholar]
- Gladstone DE, Blackford A, Cho E, Swinnen L, Kasamon Y, Gocke CD, et al. The importance of IGHV mutational status in del(11q) and del(17p) chronic lymphocytic leukemia. Clin Lymphoma Myeloma Leuk. 2012;12:132–137. doi: 10.1016/j.clml.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger JA. Chemokines and chemokine receptors in chronic lymphocytic leukemia (CLL): from understanding the basics towards therapeutic targeting. Semin Cancer Biol. 2010;20:424–430. doi: 10.1016/j.semcancer.2010.09.005. [DOI] [PubMed] [Google Scholar]
- O'Brien S, Furman RR, Coutre SE, Sharman JP, Burger JA, Blum KA, et al. Ibrutinib as initial therapy for elderly patients with chronic lymphocytic leukaemia or small lymphocytic lymphoma: an open-label, multicentre, phase 1b/2 trial. Lancet Oncol. 2014;15:48–58. doi: 10.1016/S1470-2045(13)70513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majid A, Lin TT, Best G, Fishlock K, Hewamana S, Pratt G, et al. CD49d is an independent prognostic marker that is associated with CXCR4 expression in CLL. Leuk Res. 2011;35:750–756. doi: 10.1016/j.leukres.2010.10.022. [DOI] [PubMed] [Google Scholar]
- Tobin G, Soderberg O, Thunberg U, Rosenquist R. V(H)3-21 gene usage in chronic lymphocytic leukemia—characterization of a new subgroup with distinct molecular features and poor survival. Leuk Lymphoma. 2004;45:221–228. doi: 10.1080/1042819031000147018. [DOI] [PubMed] [Google Scholar]
- Tobin G, Thunberg U, Johnson A, Thorn I, Soderberg O, Hultdin M, et al. Somatically mutated Ig V(H)3-21 genes characterize a new subset of chronic lymphocytic leukemia. Blood. 2002;99:2262–2264. doi: 10.1182/blood.v99.6.2262. [DOI] [PubMed] [Google Scholar]
- Bulian P, Shanafelt TD, Fegan C, Zucchetto A, Cro L, Nückel H, et al. CD49d is the strongest flow cytometry-based predictor of overall survival in chronic lymphocytic leukemia. J Clin Oncol. 2014;32:897–904. doi: 10.1200/JCO.2013.50.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponader S, Chen SS, Buggy JJ, Balakrishnan K, Gandhi V, Wierda WG, et al. The Bruton tyrosine kinase inhibitor PCI-32765 thwarts chronic lymphocytic leukemia cell survival and tissue homing in vitro and in vivo. Blood. 2012;119:1182–1189. doi: 10.1182/blood-2011-10-386417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsby E, Buggins A, Devereux S, Jones C, Pratt G, Brennan P, et al. Development and characterization of a physiologically relevant model of lymphocyte migration in chronic lymphocytic leukemia. Blood. 2014;123:3607–3617. doi: 10.1182/blood-2013-12-544569. [DOI] [PubMed] [Google Scholar]
- Robak T, Robak P. BCR signaling in chronic lymphocytic leukemia and related inhibitors currently in clinical studies. Int Rev Immunol. 2013;32:358–376. doi: 10.3109/08830185.2013.786711. [DOI] [PubMed] [Google Scholar]
- Barrientos J, Rai K. Ibrutinib: a novel Bruton's tyrosine kinase inhibitor with outstanding responses in patients with chronic lymphocytic leukemia. Leuk Lymphoma. 2013;54:1817–1820. doi: 10.3109/10428194.2013.796049. [DOI] [PubMed] [Google Scholar]
- Byrd JC, Furman RR, Coutre SE, Flinn IW, Burger JA, Blum KA, et al. Targeting BTK with ibrutinib in relapsed chronic lymphocytic leukemia. N Engl J Med. 2013;369:32–42. doi: 10.1056/NEJMoa1215637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.