Table 2. Thermal unfolding of apoflavodoxin variants: 3M (WT plus E20K/E72K/D126K) and mutants thereof.
| Global spectroscopic three-state fit | DSC two-state fit | |||||||
|---|---|---|---|---|---|---|---|---|
| Protein variant | ΔHNI (kcal/mol) | TmNI (°C) | ΔHIU (kcal/mol) | TmIU (°C) | ΔHND (kcal/mol) | TmND (°C) | ΔHVHa (kcal/mol) | 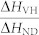 |
| 3Mb | 38.3 ± 2.8 | 50.0 ± 0.4 | 71.9 ± 2.4 | 66.7 ± 0.2 | 83.4 ± 0.2 | 67.7 ± 0.1 | 70.9 ± 0.3 | 0.85 |
| 3M/I59A | 37.2 ± 1.1 | 57.6 ± 0.2 | 87.6 ± 2.4 | 68.0 ± 0.2 | - | - | ||
| 3M/E61K | 40.9 ± 0.2 | 55.0 ± 0.1 | 83.6 ± 2.3 | 67.3 ± 0.8 | - | - | ||
| 3M/G87V | 40.1 ± 1.2 | 58.8 ± 0.2 | 80.2 ± 0.4 | 68.8 ± 0.1 | - | - | ||
| 3M/I92A | 34.2 ± 2.0 | 50.6 ± 3.1 | 65.8 ± 0.7 | 65.1 ± 1.6 | - | - | ||
| 3M/Q99A | 47.9 ± 0.6 | 58.3 ± 0.1 | 81.9 ± 1.8 | 68.6 ± 0.1 | - | - | ||
| 3M/D100N | 49.1 ± 3.2 | 68.0 ± 0.1 | 95.9 ± 1.7 | 69.7 ± 1.1 | 71.4 ± 0.2 | 69.2 ± 0.1 | 67.0 ± 0.3 | 0.94 |
| 3M/S121P | 40.8 ± 0.1 | 56.2 ± 0.1 | 75.3 ± 1.1 | 68.6 ± 0.1 | - | - | ||
| 3M/D129A | 55.1 ± 0.9 | 60.7 ± 0.3 | 83.4 ± 1.1 | 68.1 ± 0.1 | - | - | ||
| 3M/A142V | 19.7 ± 2.1 | 66.0 ± 4.7 | 83.7 ± 2.8 | 68.2 ± 0.4 | 72.1 ± 0.2 | 69.4 ± 0.1 | 75.4 ± 0.2 | 1.05 |
| 3M/I92A/I59A | 46.5 ± 1.8 | 63.9 ± 0.3 | 73.7 ± 2.0 | 69.9 ± 0.8 | - | - | ||
| 3M/I92A/G87V | 52.4 ± 6.6 | 58.9 ± 1.8 | 37.1 ± 3.3 | 75.1 ± 9.5 | - | - | ||
| 3M/I92A/D100N | 46.5 ± 2.2 | 63.2 ± 0.6 | 96.9 ± 6.2 | 69.3 ± 0.5 | - | - | ||
| 3M/I92A/A142V | 43.6 ± 2.9 | 59.0 ± 0.8 | 56.5 ± 6.9 | 67.3 ± 1.9 | - | - | ||
| 3M/A142V/G87V | - | - | - | - | 76.7 ± 0.1 | 74.4 ± 0.1 | 79.8 ± 0.2 | 1.04 |
| 3M/A142V/D100N | - | - | - | - | 82.0 ± 0.1 | 70.4 ± 0.1 | 83.6 ± 0.2 | 1.02 |
| 3M/A142V/G87V/I92A | - | - | - | - | 79.1 ± 0.1 | 74.9 ± 0.1 | 75.9 ± 0.2 | 0.96 |
| 3M/A142V/I92A/D100N | - | - | - | - | 82.3 ± 0.1 | 70.6 ± 0.1 | 85.6 ± 0.2 | 1.04 |
| 3M/A142V/I92A/I59A | - | - | - | - | 87.6 ± 0.1 | 70.3 ± 0.1 | 92.9 ± 0.2 | 1.06 |
| 3M/A142V/G87V/D100N | - | - | - | - | 84.6 ± 0.1 | 74.9 ± 0.1 | 84.5 ± 0.2 | 1.00 |
| WTb | 84.4 ± 0.2 | 55.8 ± 0.1 | 51.5 ± 0.2 | 0.61 | ||||
aVan't Hoff enthalpies, ΔHVH, have been determined by DSC, assuming a two-state protein unfolding mechanism, using the known equation: 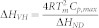 , where Tm is the mid-transition temperature, Cp,max is the maximal value of the excess molar unfolding heat capacity, and ΔHND is the calorimetric enthalpy. If the assumed two-state unfolding model is correct the calorimetric enthalpy, ΔHND, and the van't Hoff enthalpy, ΔHVH are equal (i.e. ΔHVH/ΔHND = 1.0). Otherwise, both enthalpies are significantly different.
, where Tm is the mid-transition temperature, Cp,max is the maximal value of the excess molar unfolding heat capacity, and ΔHND is the calorimetric enthalpy. If the assumed two-state unfolding model is correct the calorimetric enthalpy, ΔHND, and the van't Hoff enthalpy, ΔHVH are equal (i.e. ΔHVH/ΔHND = 1.0). Otherwise, both enthalpies are significantly different.
bThe ΔHVH/ΔHND ratio of mutant 3M indicates its thermal unfolding is not two-state. The same applies to the WT protein included in this Table for comparison.
