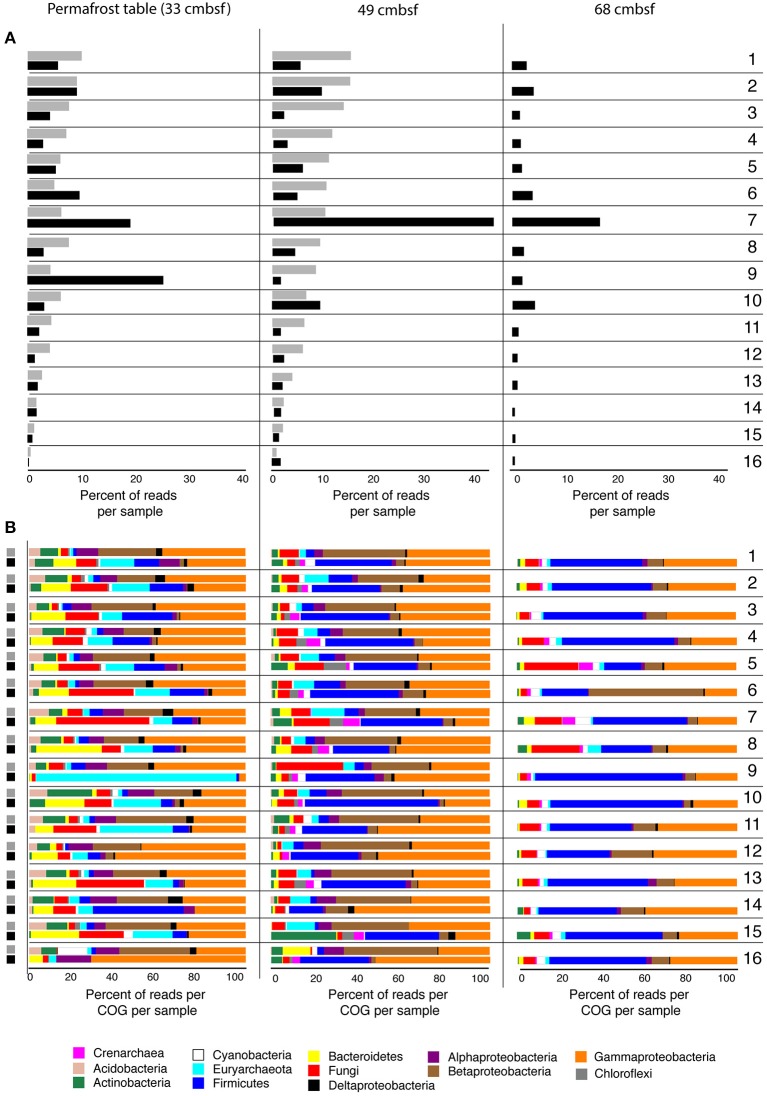
Figure 2.
(A) Relative abundance of reads assigned to COGs in metatranscriptomes from frozen (gray bars) permafrost soils of core TL08S1C3 at 33.5 ± 2 cm (permafrost table) and 49 ± 2 cm depth and at 33.5 ± 2, 49 ± 2, and 68 ± 2 cm after 11 days of incubation at 4°C (black bars). Only the 16 most (relative) abundant COGs are shown (excluding [R] General function prediction and [S] Function unknown). COGs detected in low abundance, but not shown, are [A] RNA processing and modification; [V] Defense mechanisms; [Z] Cytoskeleton. (B) The prokaryotic and fungal ORFs as percent of reads per COG per sample with homology to the following COG families: 1, Amino acid transport and metabolism; 2, Energy production and conversion; 3, Inorganic ion transport and metabolism; 4, Replication, recombination and repair; 5, Transcription; 6, Posttranslational modification, protein turnover, chaperones; 7, Translation, ribosomal structure and biogenesis; 8, Cell wall/membrane/envelope biogenesis; 9, Coenzyme transport and metabolism; 10, Carbohydrate transport and metabolism; 11, Signal transduction mechanisms; 12, Lipid transport and metabolism; 13, Nucleotide transport and metabolism; 14, Intracellular trafficking, secretion, and vesicular transport; 15, Cell cycle control, cell division, chromosome partitioning; 16, Cell motility.