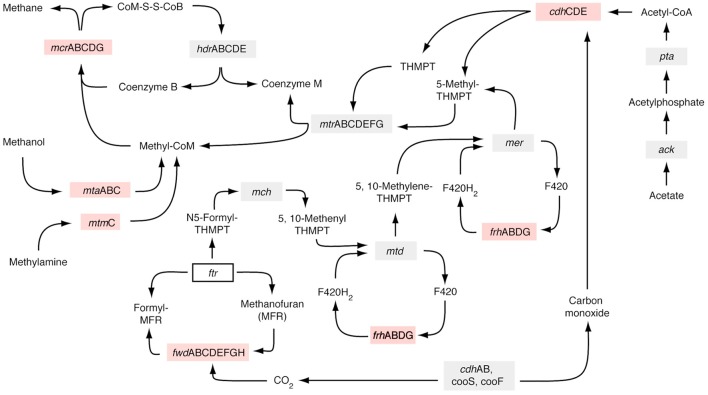
Figure 5.
Metabolic pathways actively expressed by the methanogen Methanosarcina barkeri in metatranscriptomes from frozen and thawed permafrost samples. The M. barkeri genome was chosen as a reference because it was found to be the dominant methanogen expressing mcrA after thaw (Figure 4B). Red boxes represent expressed genes that were only detected after thaw, gray boxes represent genes expressed before and after thaw, and clear boxes are genes that were not detected under any conditions. CoM-S-S-CoB, Coenzyme M 7-mercaptoheptanoylthreonine-phosphate heterodisulfide; THMPT, Tetrahydromethanopterin; hcr, Heterodisulfide reductase subunit A; mcr, Methyl-coenzyme reductase; mtr, Tetrahydromethanopterin S-methyltransferase; CoM, Coenzyme M; mtm, Monomethylamine corrinoid protein; mta Methanol-specific corrinoid protein; cdh; Acetyl-CoA decarbonylase/synthase complex; frh, Coenzyme F420 hydrogenase; mer, 5,10-methylenetetrahydromethanopterin reductase; mch, Methenyltetrahydromethanopterin cyclohydrolase; mtd, Methylenetetrahydromethanopterin dehydrogenase; fwd, Formylmethanofuran dehydrogenase; pta, Phosphate acetyltransferase; ack, Acetate kinase; cooS/F, Carbon monoxide dehydrogenase.