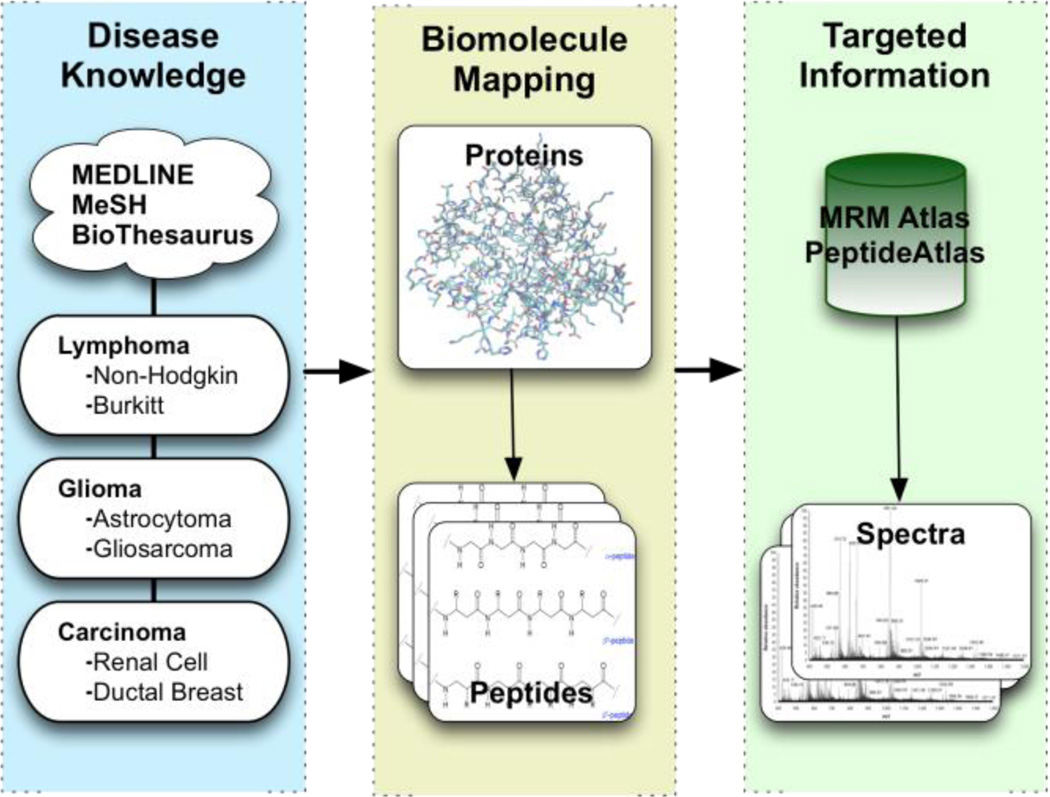
Figure 2.
We used BioThesaurus, MEDLINE, and MeSH to construct the NMD and MEDLINE Data Stores. A web service on top of the Atlas operates on the caBIG Cancer Biomedical Informatics Grid [10], and provides information about observed spectra. A data service is made available as a Google Data Source, and can be queried using GQL. This service is a read-only SQL-like interface that allows complex queries across the collected data stores for the retrieval of disease and protein related information.