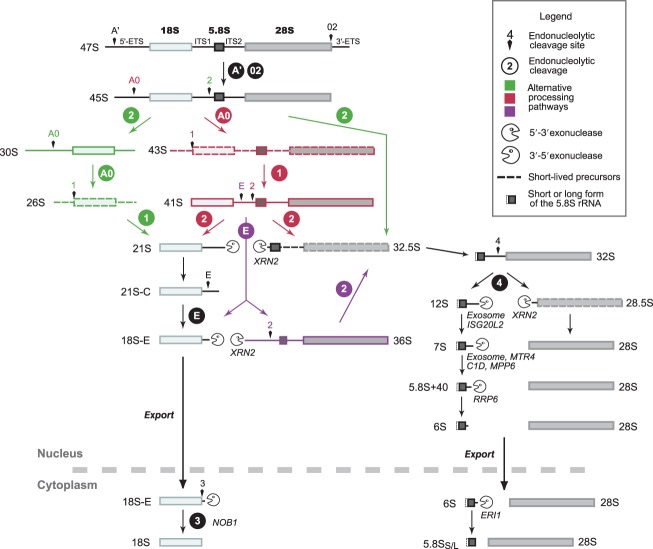
Figure 2.
Pre-ribosomal RNA (rRNA) processing in mammalian cells. The pre-rRNA processing presented here combines data from studies in human and murine cells. The nomenclature refers to human cells15,16; the corresponding nomenclature in mouse is indicated in Figure 3. Alternative cleavage sequences are depicted in different colors. Short-lived precursors are represented with dotted lines. Cleavage of the 45S pre-rRNA can either start in the 5′-ETS (red) or in the ITS1 (green), which defines two pathways. If cleavage of the 5′-ETS occurs first (41S pre-rRNA), subsequent cleavage in the ITS1 takes place either at site 2 or at site E (purple). Initial cleavage at site 2 is the major pathway in HeLa cells, considering the abundance of the 30S pre-rRNA relative to the 41S. In mouse cell lines, the 36S pre-rRNA is readily detected. The endonuclease NOB1 is necessary for maturation of the 3′ end of the 18S-E pre-rRNA in the cytoplasm. Formation of the long and short 5′ ends of the 5.8S rRNA is not fully documented in mammalian cells. The 5.8S rRNA 3′-end maturation pathway primarily involves exonucleases,16–20 but the 7S pre-rRNA was also proposed to result from endonucleolytic cleavage of the 12S pre-rRNA (site 4a in mouse, see Figure 3).21,22 It has not been formally demonstrated that final maturation of the 6S pre-rRNA takes place in the cytoplasm in mammalian cells, but this was shown in Xenopus laevis23 and Saccharomyces cerevisiae.14