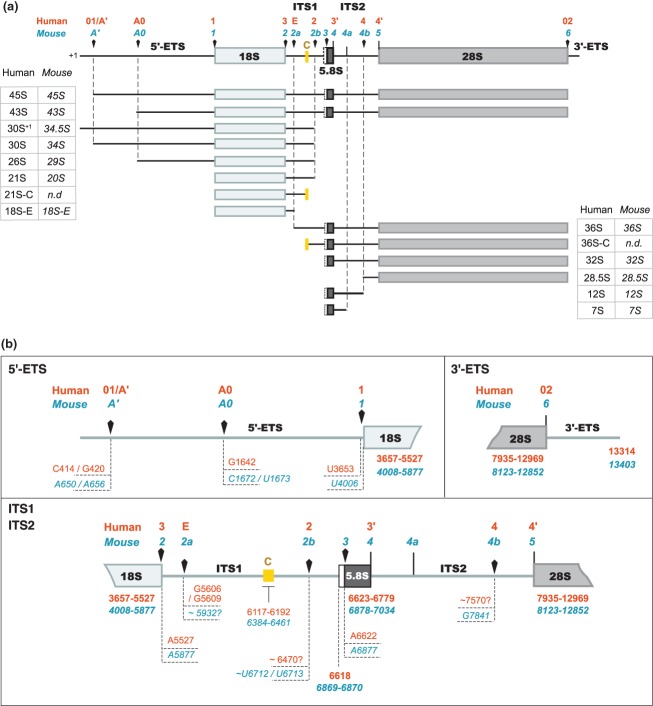
Figure 3.
The different pre-rRNAs detected in human and murine cells and their nomenclature.15,16,24,39,40,44,47,59,66 (a) Definition of the major pre-ribosomal RNAs (rRNAs). Arrowheads indicate the endonucleolytic cleavage sites. The yellow box corresponds to a highly conserved domain in ITS1 among mammals. Its boundaries define the 5′ end of the 36S-C and the 3′ end of the 21S-C, suggesting that exonucleolytic trimming of these extremities is stopped by a secondary structure and/or a protein complex anchored on this region.16 The 36S-C and the 30S+1/34.5S species are examples of precursors that are only detected upon perturbation of ribosome biogenesis. (b) Map of the processing sites in the human (orange) and murine (blue) pre-rRNAs. Arrowheads indicate the endonucleolytic cleavage sites. The nucleotides correspond to the residue located 5′ to the cleavage site. The extremities of the 18S, 5.8S, and 28S rRNAs are indicated in bold letters. The mapping of these sites is partly discussed in the text and was extensively reviewed by Mullineux and Lafontaine.59 Human site E was determined by primer extension and 3′-RACE.16 Localization of site 4b in mouse was mapped by primer extension.44 The human sites equivalent to sites 3 and 4a in mouse have not been formally designated. Details on site 4a are given in the text. The numbering of the nucleotides refers to GenBank sequences U13369.1 (human rDNA) and BK000964.3 (mouse rDNA).