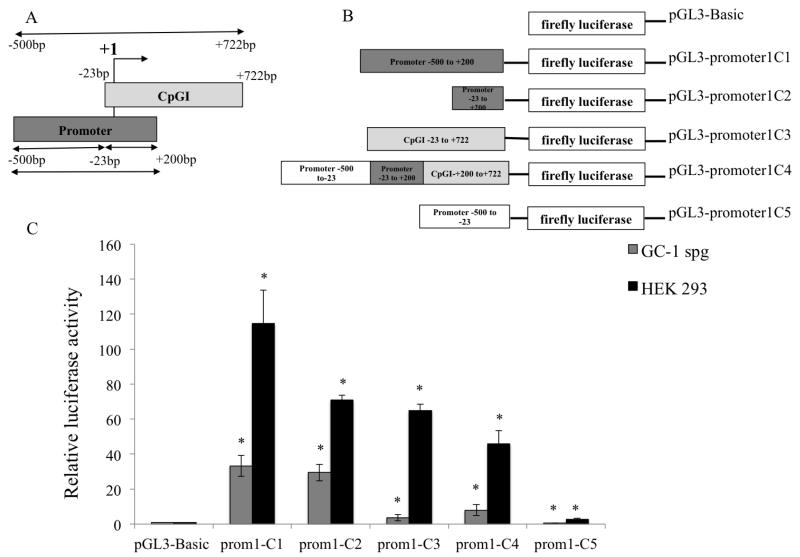
Figure 3. Functional characterization of the putative SLC9B1 promoter 1 in HEK 293 and GC-1spg cells.
(A) Schematic representation of the hNHEDC1 P1 and hNHEDC1 CpGI is shown with respect to the TSS (+1). The region including the entire CpGI extends from −500bp to +722bp relative to the TSS. P1 and CpGI overlap at −23bp to +200bp. (B) P1 was cloned upstream of the firefly luciferase gene to generate the pGL3-promoter1C1 construct. Additionally, three constructs namely: pGL3-promoter1C3, pGL3-promoter1C4 and pGL3-promoter1C5 were generated which include just the CpGI (−23bp to +722bp), P1 with the full length CpGI (−500bp to +722bp) and P1 excluding the CpGI (−500bp to −23bp) respectively. The pGL3-promoter1C2 construct contains the −23bp to +200bp overlapping region. (C) All the reporter constructs generated were tested for the ability to drive firefly luciferase expression in HEK 293 and GC-1spg cells. The reporter activity is shown as the ratio of firefly luminescence of the pGL3-hNHEDC1 promoter vectors to the pGL3-Basic (empty vector) each normalized to the Renilla luciferase luminescence and is depicted as fold increase over the promoterless pGL3-Basic vector activity (set at 1.0). Bars indicate mean±SD of three independent transfections performed in duplicate. (*) indicates statistically significant difference with p values < 0.05. Note: prom1 indicates the SLC9B1 promoter 1.