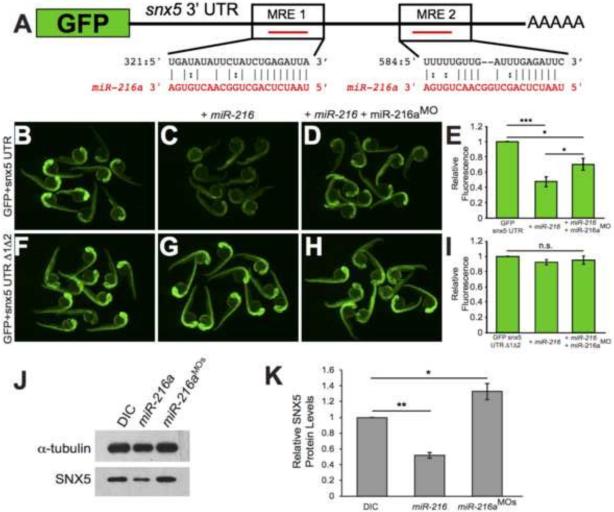
Figure 2. snx5 is a target of miR-216a.
The coding sequence of GFP was fused to the 3’UTR of snx5. Predicted pairing of each MRE in the 3’ UTR (black) and miR-216a (red) are pictured. (B) Embryos injected at the 1-cell stage with GFP-snx5 3’ UTR reporter mRNA alone, with miR-216a (C), or with miR-216a and miR-216aMO (D) were imaged using a fluorescence dissecting scope at 1 dpf. (F) Both MREs were deleted from the GFP-snx5 3’ UTR reporter. Embryos injected at the 1-cell stage with this mRNA alone, with miR-216a (G), or with miR-216a and miR-216aMO (H) were imaged at 1 dpf using a fluorescence dissecting scope. (E, I) Relative fluorescence was quantified using ImageJ, and comparisons were made using one-way ANOVA with Bonferroni's correction. (J) Western blots for SNX5 and alpha-tubulin were performed on protein lysates from 1 dpf zebrafish injected at 1-cell stage with dye control (DIC), miR-216a, or miR-216aMOs. (K) Band density was quantified using ImageJ, and comparisons were made using one-way ANOVA with Bonferroni's correction. *, p<0.05; **, p<0.01; ***, p<0.001. Error bars show SEM.)