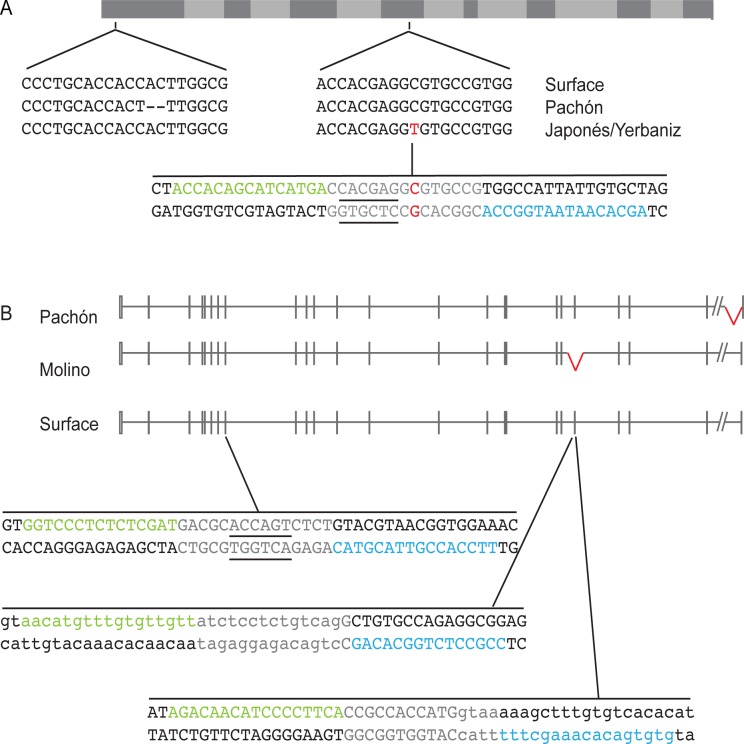
Fig 1. mc1r and oca2 TALEN targeting design.
A. Diagram of the mc1r annotated coding sequence with the location and sequences of the Pachón, Japonés and Yerbaniz mutations highlighted. The transmembrane domains are indicated in light gray. The two base pair deletion in Pachón is indicated by dashed lines. The single nucleotide change in the Japonés and Yerbaniz populations is in red. The sequence targeted by the TALEN is indicated below. The TALEN 1 binding site is in green and the TALEN 2 binding site is in blue. The spacer region is gray. The underlined sequence is the BssSI restriction enzyme recognition sequence used for genotyping. The gene structure is based on the Astyanax genome sequence database and Gross et al. 2009 [19].
B. Diagram of the oca2 gene. Boxes indicate exons and lines indicate introns. The empty boxes are UTR and the closed boxes are coding sequence. The slanted lines indicate a region of the genome left out because the distance is unknown. Note that the small scale of the figure resulted in some exons not being to scale and the size of some introns being so small that they appear to be one continuous exon. The exon 24 Pachón and the exon 21 Molino deletions are indicated in red. The amount of intronic sequence deleted in these populations is currently unknown. TALENs were designed targeting exon 9 and the either end of exon 21. The TALEN 1 binding sites are in green and the TALEN 2 binding sites are in blue. The spacer regions are gray. The underlined sequence in the exon 9 TALEN is the BsrI restriction enzyme recognition sequence used for genotyping. The exon sequence is capitalized and the intronic sequence in the two exon 21 TALENs is lower case. The genomic structure is based on the Astyanax genome sequence database and Protas et al. 2006 [17]. Both gene structures were generated using http://wormweb.org/exonintron and then modified.