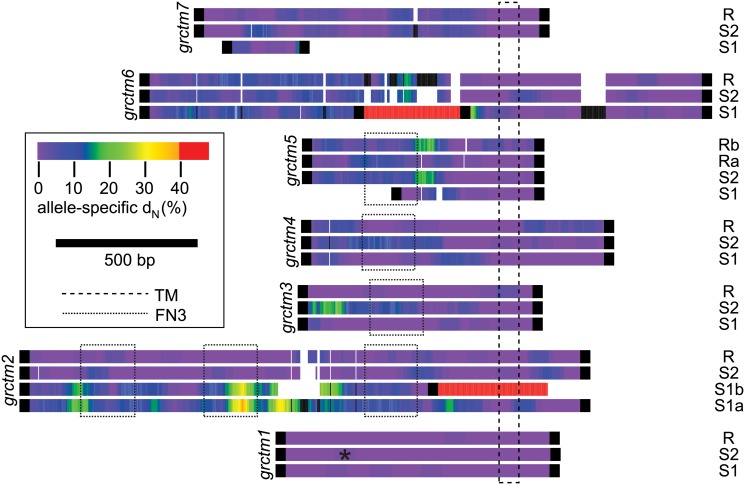
Fig 4. Allele-specific nonsynonymous substitution among alleles of the seven TM1 genes.
Alleles of the same gene are aligned to each other (gaps indicated by whitespace), but across genes only the transmembrane domain (TM) is aligned (extracellular (N-terminal) regions are to the left of the TM). If a haplotype includes two copies of a gene that is single-copy on the other haplotypes, both copies are shown (e.g. gene grctm5 is duplicated on the R haplotype, so we label those sequences Ra and Rb). For each allele, we calculated allele-specific nonsynonymous substitution (i.e. divergence from the inferred ancestral sequence) in 75bp sliding windows, indicated by color (“allele-specific dN”). Regions with no sequence similarity are indicated in red. Substitution across a 75bp window could not be calculated in black sections. Fibronectin III domains (FN3) are shown. The premature stop codon in grctm1 S2 is shown with an asterisk. Nonsynonymous substitution is extremely high across the TM1 genes, exceeding 15% in some windows for all genes except grctm1, and occasionally reaching over 30%. Only grctm5 and grctm6 show high nonsynonymous substitution specific to R alleles.