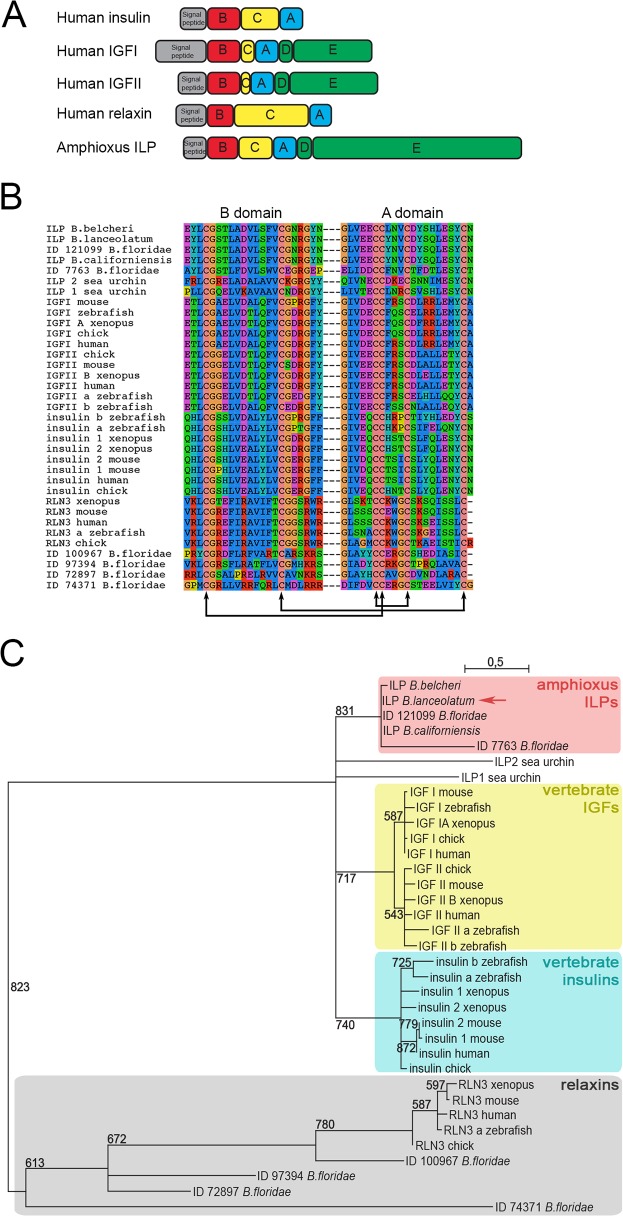
Fig 1. Structural comparison and phylogenetic relationships between amphioxus ILP and insulin-relaxin superfamily proteins in vertebrates.
A. Comparison of amphioxus ILP domain structure with human insulin, IGFs and relaxin. B. Sequence alignment of the conserved A and B domains of ILP sequences from different amphioxus species and insulin, IGF and relaxin sequences from vertebrates and sea urchin. Black arrows indicate the conserved cysteine residues necessary for the formation of disulfide bonds during the processing of precursor proteins. C. Phylogenetic maximum likelihood analysis of chordate insulin/IGF subfamily members. The red square highlights amphioxus ILP sequences, the blue square highlights vertebrates insulin sequences and the yellow square highlights vertebrates IGF sequences; the grey square indicates the outgroup made with relaxin sequences. B. lanceolatum ILP sequence identified in this study is pointed with the red arrow. Bootstrap values derived from 1000 runs are shown. Branches with bootstraps lower than 500 were collapsed. The scale bar indicates the average number of amino acid substitutions per site.