Abstract
Background:
Pseudomonas aeruginosa is an opportunistic human pathogen that causes serious problems, especially in people, who have immunodeficiency. In recent times, metallo-β-lactamase (MBLs) resistance in this bacterium has led to some difficulties in treating bacterial infections. The metallo-beta-lactamase family of genes, including blaVIM-1, is being reported with increasing frequency worldwide. The aim of this study is the detection of the metallo-β-lactamase gene blaVIM-1 in imipenem-resistant P. aeruginosa (IRPA) strains isolated from hospitalized patients.
Materials and Methods:
In this study, 106 P. aeruginosa samples were isolated from various nosocomial infections. The isolates were identified, tested for susceptibility to various antimicrobial agents by the Kirby-Bauer disk diffusion method, and all the imipenem-resistant isolates were screened for the presence of MBLs by using the combined disk (IMP-EDTA). The minimal inhibitory concentration (MIC) of imipenem was determined by E-test on the Mueller-Hinton agar. To detect the blaVIM-1 gene, the isolates were subjected to a polymerase chain reaction (PCR).
Results:
Of all the P. aeruginosa isolates, 62 (58.5%) were found to be imipenem-resistant P. aeruginosa (MIC ≥32 μg/ml). Twenty-six (42%) of the imipenem-resistant isolates were MBL positive. None of these isolates carried the blaVIM-1 gene using the PCR assay.
Conclusion:
The results demonstrated the serious therapeutic threat of the MBL-producing P. aeruginosa populations. The rate of imipenem resistance due to MBL was increased dramatically. Early detection and infection-control practices are the best antimicrobial strategies for this organism. None of MBL-producing isolates in this study carry the blaVIM-1 gene; therefore, another gene in the MBL family should be investigated.
Keywords: blaVIM-1, imipenem, metallo-β-lactamase, Pseudomonas aeruginosa
INTRODUCTION
Pseudomonas aeruginosa is one of the important pathogens most frequently responsible for nosocomial infections and it is an opportunistic human pathogen.[1] This pathogen is the major cause of morbidity and mortality in immunocompromised patients such as cystic fibrosis, burn issues, cancer, and patients in Intensive Care Units.[2,3,4,5] They exhibit intrinsic resistance to several antimicrobial agents, including most b-lactams, and they often carry acquired resistance determinants that further reduce their spectrum of susceptibility.[6,7,8] Antimicrobial resistance in this species is a problem of growing concern and limits our therapeutic alternatives. Carbapenems are commonly used as last-resort drugs for the treatment of infections caused by multidrug-resistant P. aeruginosa isolates. However, intensive use of carbapenems in the treatment of nosocomial P. aeruginosa infections has facilitated the emergence of mechanisms that confer resistance to carbapenems, such as, diminished permeability, overexpression of the intrinsic efflux systems, and production of carbapenemases.[9] Acquisition of class B metallo-b-lactamases (MBLs) constitute a growing family of carbapenem-hydrolyzing beta-lactamases, among P. aeruginosa strains.[10] These enzymes efficiently hydrolyze all beta lactam compounds, except aztreonam.[11] These enzymes belong to Ambler class B and Bush group 3 and require divalent cations, usually zinc (Zn), as a cofactor for enzyme activity. They are inhibited by metal chelators, such as, Ethylenediaminetetraacetic acid (EDTA), but are not affected by therapeutic beta-lactamase inhibitors like sulbactam, tazobactam or clavulanic acid.[12] Metallo-β-lactam genes are usually a part of an integron structure and are carried on transferable plasmids, but can also be part of the chromosome. On account of integron-associated gene cassettes, MBL-producing P. aeruginosa isolates are often resistant to different groups of antimicrobial agents, which can be transferred to various types of bacteria.[13] Therefore, MBL-producing strains are important from an infection-control perspective. The MBLs are divided into the following six groups based on molecular structure: IMP, VIM, SIM, SPM, GIM, and AIM.[14] The the three main clusters of the VIM MBL, among 11 variants of this type, have been identified so far, represented by blaVIM-1, VIM-2, and VIM-7.[15] Metallo-β-lactamases producing P. aeruginosa isolates were first reported in Japan in 1991, and since then have been detected in various countries.[16] The present study was completed to detect blaVIM-1 producing P. aeruginosa, isolated from patients admitted to the hospitals of Isfahan.
MATERIALS AND METHODS
One hundred and six isolates of P. aeruginosa were collected from the hospitalized patients in Isfahan city of Iran, between October 2012 and November 2013. These strains were separated from blood, wound, sputum, urine, eye infection, catheters, ear, peritoneum, and burning specimen Only one isolate per patient was included in the study. These isolates were identified as P. aeruginosa based on the colonial morphology on Sytrymyd agar and Pseudomonas agar, Gram stain characteristics, Oxidative-fermentative test, oxidase test, growth in 42°C, and pigment production tests. Strains were preserved in Brain–Heart infusion broth (Merck-German) containing 20% glycerol.[17]
Antibiotic susceptibility tests
The antibiotic susceptibility test was done by the disk diffusion method (Kirby-Bauer) on Muller-Hinton agar plates (Merck-German) according to the CLSI (Clinical and Laboratory Standards Institute) guidelines. The antimicrobial disks used were amikacin (10 μg), ciprofloxacin (10 μg), ceftazidime (10 μg), gentamicin (10 μg), imipenem (10 μg), meropenem (10 μg), cefotaxime (10 μg), cefepime (10 μg), and aztreonam (10 μg), which were obtained from MAST (Merseyside, UK). P. aeruginosa ATCC 27853 were used as control for, susceptibility testing. The bacteria were inoculated into BHI, and incubated at 35°C for two-to-four hours until it reached the turbidity of a 0.5 McFarland standard. Then using a sterile swab they were cultured on Muller-Hinton agar plate. Then, antimicrobial disks were placed on the plate and incubated at 37°C for 18 to 24 hours. After incubation, the diameters of the zone of complete inhibition were measured.[18,19]
Minimum inhibitory concentration
Minimum inhibitory concentration (MIC) test was performed by the E-test method for imipenem-resistant strains. Mueller-Hinton agar plates, the recommended medium, were streaked by using cotton swabs. The E-test strips were impregnated with imipenem and then applied on the plates, which were incubated at 35°C in air for 16 to 20 hours.[20] P. aeruginosa strains with MIC ≥8 μg/ml to imipenem are referred to as resistant.[21]
Ethylenediaminetetraacetic-imipenem test
Phenotypic detection of MBL production (EDTA-IMP) was carried out for imipenem-resistant isolates (by dissolving 186.1 g of disodium EDTA in 2H2O in 1000 ml of distilled water, 0.5 M EDTA solution was provided and adjusted to pH 8 by adding NaOH. Next, 750 μg of prepared solution was added to the imipenem disk. The EDTA-imipenem disk plus the imipenem disk were placed in a plate containing Muller-Hinton agar and cultured P. aeruginosa. After 18–24 hours of incubation at 37°C an organism was considered MBL positive if the inhibition zone diameter increased by 7 mm or more toward the IMP + EDTA, in comparison to the IMP disk alone.[22]
Molecular analysis
Total DNA from the P. aeruginosa strains was extracted by the boiling method; this caused the cell wall to denaturate and lysis.[23] PCR assay was performed to detect the blaVIM-1 gene. A strain of P. aeruginosa known to produce VIM-1 metallo-beta-lactamase was used as a positive control in all cases.[17]
Polymer chain reaction tests were done under the following program
Polymer Chain Reaction for detection of the blaVIM-1 gene was performed on total DNA using VIM-1 primers (Forward: 5 × TTA TGG AGC AGC AAC CGA TGT 3 × and Reverse: 5 × CAA AAG TCC CGC TCC AAC GA 3×). The size of the amplification product was 830 bp.[24] The cycling parameters for the blaVIM-1 gene were: 94°C for three minutes, followed by 35 cycles of denaturation at 94°C for one minute, annealing at 55°C for one minute, extension at 72°C for two minutes, and a final extension at 72°C for seven minutes. The PCR products were loaded into a 2.0% agarose gel, stained with 1% ethidium bromide, electrophoresed, and visualized under ultraviolet (UV) light.[24] The positive controls were the MBL-producing strains. Amplification reactions were performed in a final volume of 25 μL, containing 200 μM concentrations of dNTPs (Fermentas-Canada), 10 pM of each primer (eurofins MWG/Operon, Ebersberg, Germany), 0.8 mM MgCl2 (Fermentas-Canada), 0.5 U Taq polymerase, and 50 ng DNA templates.[25]
RESULTS
Among 106 strains collected, the majority was isolated from urine (36%) and minority was related to the ear (1%) [Figure 1]. The disk diffusion method showed that 62 isolates (58.5%) were resistant to imipenem and meropenem. Other antibiotic resistances are shown in Figure 2. MIC test illustrated that MIC in all of imipenem-resistant P. aeruginosa (IRPA) strains were MIC ≥32 μg/ml [Figure 3]. In the phenotypic method, 26 (42%) imipenem-resistant isolates produced the MBL enzyme (increase of ≥7 mm in a zone diameter of EDTA-IMP disk compared to the imipenem disk) [Figure 4]. The molecular assay (PCR method) did not detect the blaVIM-1 gene in this study [Figure 5].
Figure 1.
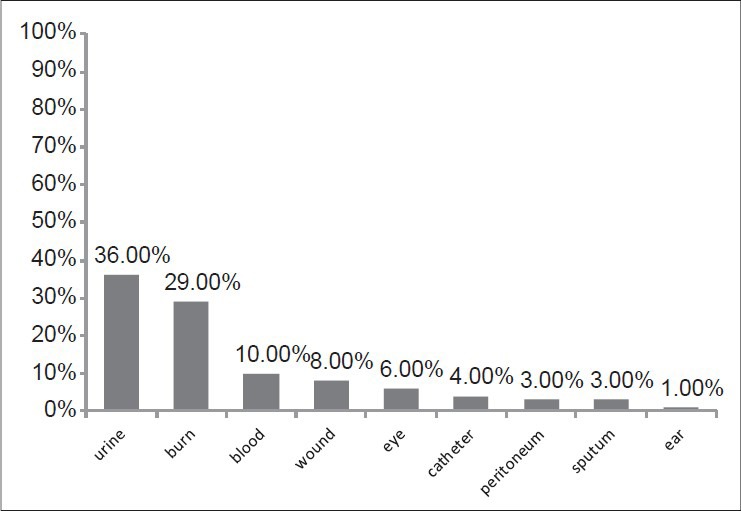
The frequency of isolation of P. aeruginosa isolated from a clinical specimen
Figure 2.
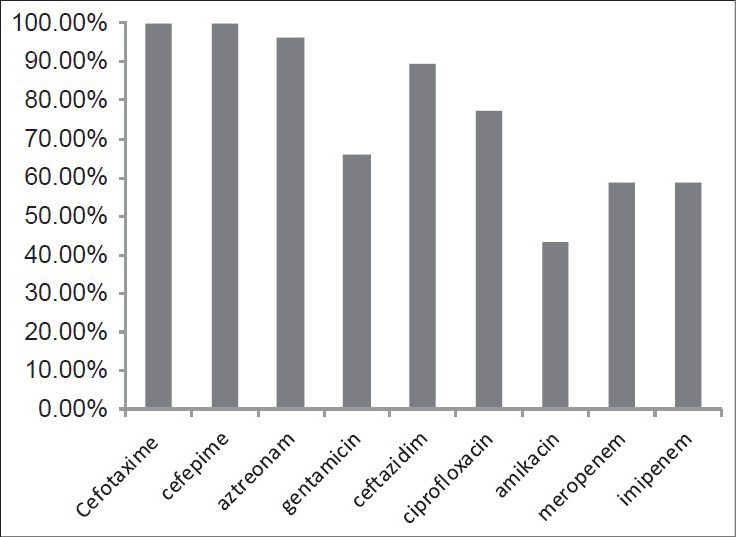
Resistance pattern of P. aeruginosa isolates to different antimicrobial agents
Figure 3.
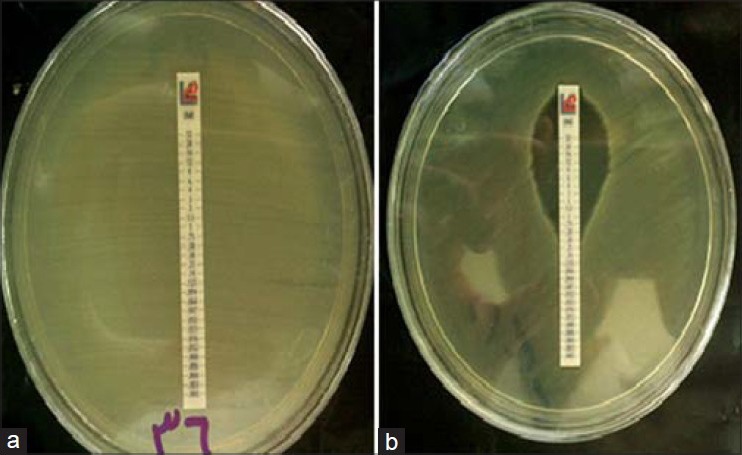
Measuring the MIC of imipenem-resistant P. aeruginosa isolates and (a) Compared with the control strain (b) by the E-test Method
Figure 4.
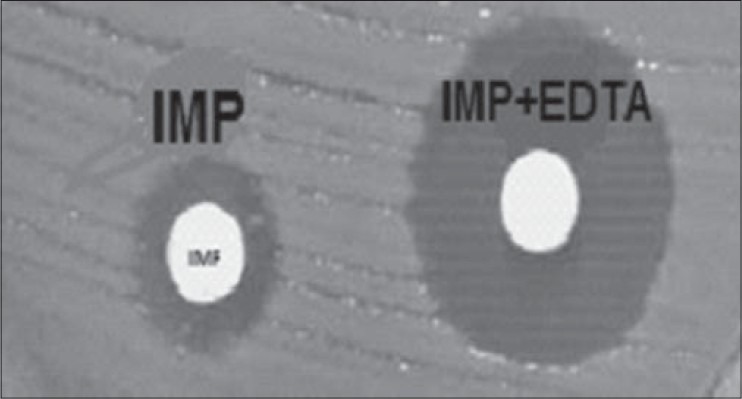
Phenotypic Detection of MBLs by Combined disk (EDTA+IMP) among the P. aeruginosa Isolates
Figure 5.
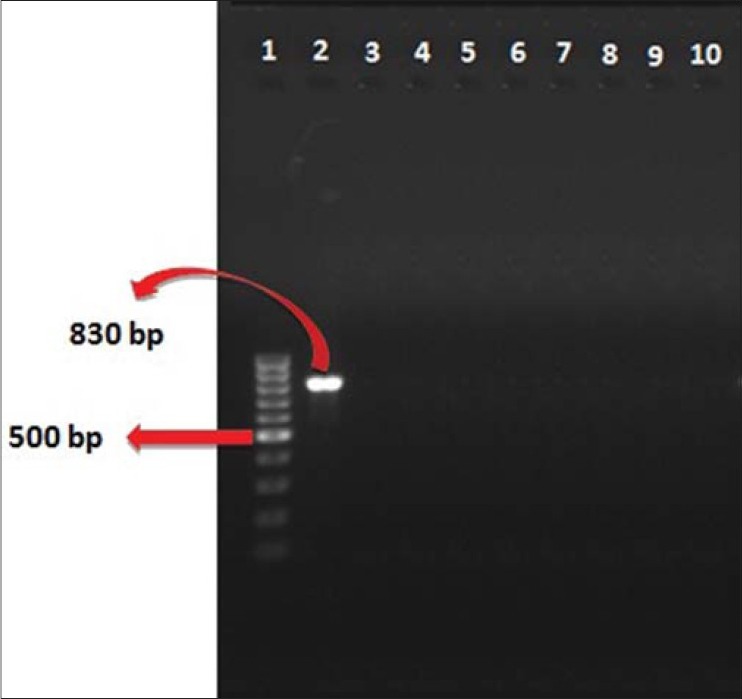
PCR assay for the Detection of the blaVIM-1 gene (product size: 830 bp) (Lane 1: DNA ladder 100 bp, Lane 2: Positive control, Lane 3: Negative control, Lanes 4-10: Amplified products)
DISCUSSION
The rapid spread of MBLs among major gram-negative pathogens, particularly P. aeruginosa, is an emerging threat and a matter of concern worldwide.[26,27] There is more concern about P. aeruginosa, which is one of the opportunistic pathogens inpatients with immune deficiency.[27,28] Imipenem-resistant P. aeruginosa (IRPA) is a current and significant concern, especially because of the limited therapeutic options for this pathogen. MBL enzymes may play a critical role in IRPA, given that there is a high possibility of these carbapenemases being spread among nosocomial isolates. The prevalence of MBLs has been increasing significantly. MBLs now account for up to 40% of the IRPA cases worldwide; furthermore, the enzyme types may vary by regional areas.[24] In this study, 108 P. aeruginosa strains were obtained from different hospitalized patients from the different hospitals of Isfahan. The antibiotic susceptibility pattern was determined in these strains and imipenem-resistant strains were subjected to be tested for MIC, EDTA-IMP, and molecular analysis. Metallo-beta-lactamases are a group of β-lactamase enzymes that have one or two zinc (Zn) in their active site to cleave the amide bond of the β-lactam ring to inactive β-lactam antibiotics. In the recent years, nosocomial outbreaks of MBL-producing bacteria have been reported worldwide.[29] The blaVIM-1 gene was first reported in P. aeruginosa in Italy.[25] As far as recent studies have shown, this enzyme has spread significantly. Furthermore, there are 20 various known varieties of blaVIM alleles all over the world.[19] This gene has been reported in different areas in Iran, but our study showed that none of IRPA strains carry blaVIM-1 in the hospitals of Isfahan.
In Italy, three studies have been done. According to Francesco luzzaro et al. (2004), the blaVIM-1 gene was detected in only one isolate from the 506 isolates of IRPA.[30] Cristiana Lagatolla, conducted two studies. In the first study on 89 IRPA isolates, 54 (84%) isolates had the blaVIM-1 gene.[31] The second study reported that 86 strains between 174 IRPA had the blaVIM-1 gene.[32] In another study in Brazil, by Fernanda et al., (2009), 31 P. aeruginosa isolates were investigated, but no blaVIM-1 gene existed in these isolates.[17] Also, Franco et al. (2010), showed that this gene did not exist in 238 isolates of P. aeruginosa in his study.[24] Two studies were conducted in Spain. According to a study by Carvalho et al. (2005), 27 isolates of P. aeruginosa were investigated, where the blaVIM-1 gene was not identified in these strains.[33] Also, in another study on 236 P. aeruginosa isolates by Gutierrez et al. (2007), the blaVIM-1 gene was not detected.[34] In 2008, Khosravi and colleagues collected 100 P. aeruginosa isolates. Disk diffusion showed that 41 of the isolates were resistant to imipenem, eight (19.51%) appeared to produce MBL, and eight imipenem-resistant strains were blaVIM-1 positive.[35] In 2010, Saderi and colleagues obtained 100 P. aeruginosa from 100 burn patients in Tehran. In that study the prevalence of MBL-producing P. aeruginosa and detection of the blaVIM-1 gene were determined. Sixty-five out of 69 imipenem-resistant P. aeruginosa showed MBL activity, while only 13 of them had the MBL gene. The blaVIM-1 was not found finally.[35] In 2012, Forozesh Fard and colleagues collected 11 P. aeruginosa isolates. All the isolates were susceptible to imipenem, and all of them were blaVIM-1 negative.[18] Compared to our study, the imipenem-resistant strains in Isfahan Hospitals showed higher resistance to imipenem (37%), and MBLs existed in a high level, but the blaVIM-1 gene had a low incidence or even maybe did not exist, which shows that another MBL genes were probably involved.
A combination therapy can be useful to prevent resistance during therapy. Regarding the horizontal transmission of integron-associated MBL genes, detecting MBL-positive strains is essential. Moreover, the invention of new methods for identifying MBL-positive bacteria, and screening involving people, must be done in hospitals regularly.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Patzer JA, Dzierzanowska D. Increase of imipenem resistance among Pseudomonas aeruginosa isolates from a polish paediatric hospital (1993-2002) Int J Antimicrob Agents. 2007;29:153–8. doi: 10.1016/j.ijantimicag.2006.08.044. [DOI] [PubMed] [Google Scholar]
- 2.Castanheira M, Toleman MA, Jones RN, Schmidt FJ, Walsh TR. Molecular characterization of a beta-lactamase gene, blaGIM-1, encoding a new subclass of metallo-beta-lactamase. Antimicrob Agents Chemother. 2004;48:4654–61. doi: 10.1128/AAC.48.12.4654-4661.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crespo MP, Woodford N, Sinclair A, Kaufmann ME, Turton J, Glover J, et al. Outbreak of carbapenem-resistant Pseudomonas aeruginosa producing VIM-8, a novel metallo-beta-lactamase, in a tertiary care center in Cali, Colombia. J Clin Microbiol. 2004;42:5094–101. doi: 10.1128/JCM.42.11.5094-5101.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farra A, Islam S, Strålfors A, Sörberg M, Wretlind B. Role of outer membrane protein OprD and penicillin-binding proteins in resistance of Pseudomonas aeruginosa to imipenem and meropenem. Int J Antimicrob Agents. 2008;31:427–33. doi: 10.1016/j.ijantimicag.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 5.Huang YT, Chang SC, Lauderdale TL, Yang AJ, Wang JT. Molecular epidemiology of carbapenem-resistant Pseudomonas aeruginosa carrying metallo-beta-lactamase genes in Taiwan. Diagn Microbiol Infect Dis. 2007;59:211–6. doi: 10.1016/j.diagmicrobio.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Nikaido H. Prevention of drug access to bacterial targets: Permeability barriers and active efflux. Science. 1994;264:382–8. doi: 10.1126/science.8153625. [DOI] [PubMed] [Google Scholar]
- 7.Nikaido H, Nikaido K, Harayama S. Identification and characterization of porins in Pseudomonas aeruginosa. J Biol Chem. 1991;266:770–9. [PubMed] [Google Scholar]
- 8.Lodge JM, Minchin SD, Piddock LJ, Bushy SJ. Cloning, sequence and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC beta-lactamase. Biochem J. 1990;272:627–31. doi: 10.1042/bj2720627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tam VH, Chang KT, LaRocco MT, Schilling AN, McCauley SK, Poole K, et al. Prevalence, mechanisms, and risk factors of carbapenem resistance in bloodstream isolates of Pseudomonas aeruginosa. Diagn Microbiol Infect Dis. 2007;58:309–14. doi: 10.1016/j.diagmicrobio.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Walsh TR. Clinically significant carbapenemases: An update. Curr Opin Infect Dis. 2008;21:367–71. doi: 10.1097/QCO.0b013e328303670b. [DOI] [PubMed] [Google Scholar]
- 11.Queenan AM, Bush K. Carbapenemases: The versatile beta-lactamases. Clin Microbiol Rev. 2007;20:440–58. doi: 10.1128/CMR.00001-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livermore DM, Woodford N. Carbapenemases: A problem in waiting? Curr Opin Microbiol. 2000;3:489–95. doi: 10.1016/s1369-5274(00)00128-4. [DOI] [PubMed] [Google Scholar]
- 13.Nordmann P, Poirel L. Emerging carbapenemases in Gram-negative aerobes. Clin Microbiol Infect. 2002;8:321–31. doi: 10.1046/j.1469-0691.2002.00401.x. [DOI] [PubMed] [Google Scholar]
- 14.Sacha P, Wieczorek P, Hauschild T, Zórawski M, Olszańska D, Tryniszewska E. Metallo-beta-lactamases of Pseudomonas aeruginosa - A novel mechanism resistance to beta-lactam antibiotics. Folia Histochem Cytobiol. 2008;46:137–42. doi: 10.2478/v10042-008-0020-9. [DOI] [PubMed] [Google Scholar]
- 15.Henrichfreise B, Wiegand I, Sherwood KJ, Wiedemann B. Detection of VIM-2 metallo-beta-lactamase in Pseudomonas aeruginosa from Germany. Antimicrob Agents Chemother. 2005;49:1668–9. doi: 10.1128/AAC.49.4.1668-1669.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pitout JD, Gregson DB, Poirel L, McClure JA, Le P, Church DL. Detection of Pseudomonas aeruginosa producing metallo-beta-lactamases in a large centralized laboratory. J Clin Microbiol. 2005;43:3129–35. doi: 10.1128/JCM.43.7.3129-3135.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wirth FW, Picoli SU, Cantarelli VV, Gonçalves AL, Brust FR, Santos LM, et al. Metallo-beta-lactamase-producing Pseudomonas aeruginosa in two hospitals from southern Brazil. Braz J Infect Dis. 2009;13:170–2. doi: 10.1590/s1413-86702009000300003. [DOI] [PubMed] [Google Scholar]
- 18.Forozsh FM, Irajian G, Moslehi TZ, Fazeli H, Salehi M, Rezania S. Drug resistance pattern of Pseudomonas aeruginosa strains isolated from cystic fibrosis patients at Isfahan AL Zahra hospital, Iran (2009-2010) Iran J Microbiol. 2012;4:94–7. [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta V. Metallo beta lactamases in Pseudomonas aeruginosa and Acinetobacter species. Expert Opin Investig Drugs. 2008;17:131–43. doi: 10.1517/13543784.17.2.131. [DOI] [PubMed] [Google Scholar]
- 20.Lee K, Yong D, Yum JH, Lim YS, Bolmström A, Qwärnström A, et al. Evaluation of Etest MBL for detection of blaIMP-1 and blaVIM-2 allele-positive clinical isolates of Pseudomonas spp. and Acinetobacter spp. J Clin Microbiol. 2005;43:942–4. doi: 10.1128/JCM.43.2.942-944.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.USA. Wayne, PA: CLSI; 2013. Clinical and Laboratory Standards Institute. Performance standards for Antimicrobial susceptibility Testing; Twenty-Third Informational Supplement: Document M100-S23. [Google Scholar]
- 22.Hemalatha V, Sekar U, Kamat V. Detection of metallo betalactamase producing Pseudomonas aeruginosa in hospitalized patients. Indian J Med Res. 2005;122:148–52. [PubMed] [Google Scholar]
- 23.Cornaglia G, Akova M, Amicosante G, Cantón R, Cauda R, Docquier JD, et al. ESCMID Study Group for Antimicrobial Resistance Surveillance (ESGARS).Metallo-beta-lactamases as emerging resistance determinants in Gram-negative pathogens: Open issues. Int J Antimicrob Agents. 2007;29:380–8. doi: 10.1016/j.ijantimicag.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 24.Franco MR, Caiaffa-Filho HH, Burattini MN, Rossi F. Metallo-beta-lactamases among imipenem-resistant Pseudomonas aeruginosa in a Brazilian University hospital. Clinics (Sao Paulo) 2010;65:825–9. doi: 10.1590/S1807-59322010000900002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadeghi A, Rahimi B, Shojapour M. Molecular detection of metallo-β-lactamase genes blaVIM-1, blaVIM-2, blaIMP-1, blaIMP-2 and blaSPM-1 in Pseudomonas aeruginosa isolated from hospitalized patients in Markazi province by Duplex-PCR. Afr J Microbiol Res. 2012;6:2965–9. [Google Scholar]
- 26.Nam Hee R, Jung Sook H, Dong Seok J, Jae Ryong K. Prevalence of Metallo-β-lactamases in Pseudomonas aeruginosa and Acinetobacter baumannii. Korean J Clin Microbiol. 2010;13:169–72. [Google Scholar]
- 27.Spilker T, Coenye T, Vandamme P, LiPuma JJ. PCR-based assay for differentiation of Pseudomonas aeruginosa from other Pseudomonas species recovered from cystic fibrosis patients. J Clin Microbiol. 2004;42:2074–9. doi: 10.1128/JCM.42.5.2074-2079.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng K, Smyth RL, Gvan JR, Doherty C, Winstannley C, Denning N, et al. Spread of beta-lactam resistant Pseudomonas aeruginosa in a cystic fibrosis clinic. Lancet. 1996;348:639–42. doi: 10.1016/S0140-6736(96)05169-0. [DOI] [PubMed] [Google Scholar]
- 29.Yan JJ, Hsueh PR, Ko WC, Luh KT, Tsai SH, Wu HM, et al. Metallo-beta-lactamases in clinical Pseudomonas isolates in Taiwan and identification of VIM-3, a novel variant of the VIM-2 enzyme. Antimicrob Agents Chemother. 2001;45:2224–8. doi: 10.1128/AAC.45.8.2224-2228.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lagatolla C, Tonin EA, Monti-Bragadin C, Dolzani L, Gombac F, Bearzi C, et al. Endemic carbapenem-resistant Pseudomonas aeruginosa with acquired metallo-beta-lactamase determinants in European hospital. Emerg Infect Dis. 2004;10:535–8. doi: 10.3201/eid1003.020799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lagatolla C, Edalucci E, Dolzani L, Riccio ML, De Luca F, Medessi E, et al. Molecular evolution of metallo-beta-lactamase-producing Pseudomonas aeroginosa in a nosocomial seting of high-level endemicity. J Clin Microbiol. 2006;44:2348–53. doi: 10.1128/JCM.00258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carvalho MJ, Saavedra MJ, Correia A, Castro AP, Duarte A. Metallo-beta-lactamase in clinical Pseudomonas aeruginosa isolate in a Portuguese hospital and identification of a new VIM-2 like enzyme. Microbiol and Infect. 2005;11:409. [Google Scholar]
- 33.Gutiérrez O, Juan C, Cercenado E, Navarro F, Bouza E, Coll P, et al. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob Agents Chemother. 2007;51:4329–35. doi: 10.1128/AAC.00810-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khosravi AD, Mihani F. Detection of metallo-beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients in Ahwaz, Iran. Diagn Microbiol Infect Dis. 2008;60:125–8. doi: 10.1016/j.diagmicrobio.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 35.Saderi H, Lotfalipour H, Owlia P, Salimi H. Detection of Metallo-β-Lactamase Producing Pseudomonas aeruginosa Isolated From Burn Patients in Tehran, Iran. Labmedicine. 2010;41:609–12. [Google Scholar]


