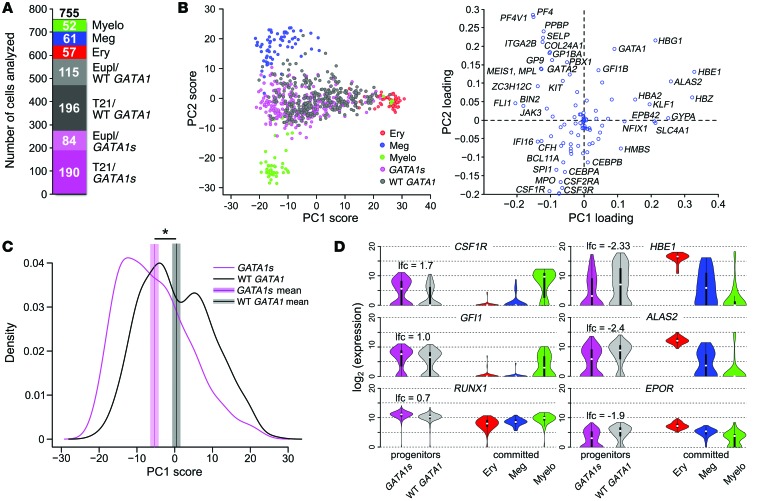
Figure 5. PCA identifies lineage signatures and reveals heterogeneity within progenitor cell populations.
Single cells within CD43+CD41+CD235+ hematopoietic progenitor populations derived from WT and mutant iPSCs were purified by flow cytometry and analyzed for gene expression by qPCR. For comparison, committed erythroid (CD41–CD235+), myeloid (CD45+CD18+), and megakaryocytic (CD41+CD42+) cells generated from WT iPSCs were purified and analyzed in parallel. (A) Numbers of analyzed single cells of each genotype (total 755 single cells). WT lineage committed cells: myeloid (myelo), megakaryocytic, and erythroid (ery). CD43+CD41+CD235+ progenitor genotypes: euploid/WT GATA1, euploid/GATA1s, T21/WT GATA1, T21/GATA1s. (B) PCA on 170 committed erythroid (red), myeloid (green), and megakaryocytic (blue) cells based on expression patterns of 94 genes (right plot). PC loadings obtained from this analysis (right) were used to project 274 GATA1s (purple) and 311 WT GATA1 (black) progenitor cells onto PC1 and PC2 (left). (C) Smooth kernel density estimate of PC1 scores of GATA1s (purple line) and WT GATA1 progenitors (black line). Purple and black vertical lines represent mean PC1 scores. Shaded areas around the mean correspond to 90% confidence interval for the mean. *P = 10–13 (Mann-Whitney U test). (D) Violin plots showing distributions of single-cell expression levels of 3 genes that were upregulated (left) and 3 genes that were downregulated (right) in GATA1s vs. WT GATA1 progenitors in 5 cell types analyzed (FDR < 0.05; Mann-Whitney U test followed by BH-FDR correction). lfc, lfc of mean expression between GATA1s and WT GATA1 progenitors.