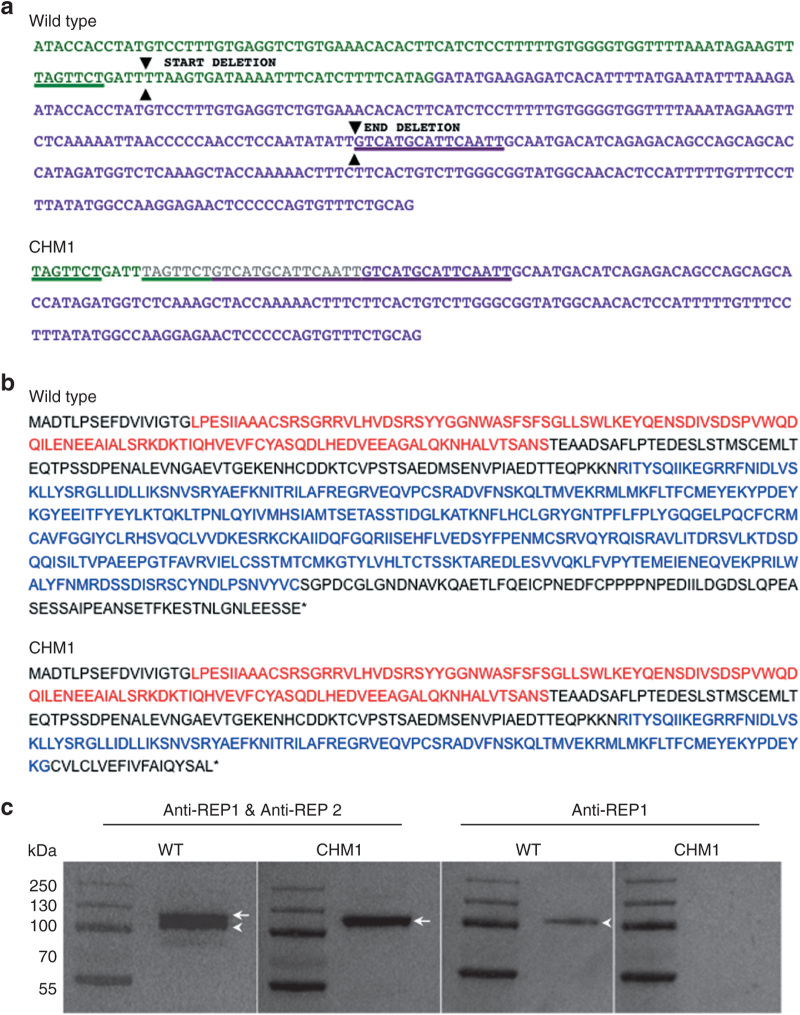
Figure 1.
Characterization of the CHM deletion of patient CHM1. (a) The sequence of exon 7 of the CHM gene is indicated in green, and the sequence of exon 8 is indicated in purple. The 7-bp sequence (underlined in green) in exon 7, and the 15-bp sequence in exon 8 (underlined in purple), which are duplicated in the DNA of CHM1 are positioned on the wild-type complementary DNA (cDNA) sequence. The limits of the resulting deletion (arrowheads) are also indicated. The duplicated and deleted cDNA sequences of patient CHM1 for the corresponding region are shown. The inserted sequence is in gray. (b) The wild-type REP1 sequence of 653 aa containing two guanosine diphosphate (GDP) dissociation inhibitor (GDI) domains, indicated in red and blue. The truncated REP1 sequence is predicted to be 332 aa long and to contain a truncated second GDI domain (in blue). (c) Western blot analysis with an antibody recognizing both REP1 and REP2 detects two bands that migrate at ~110 (arrowhead) and 120 (arrow) kDa, respectively, in wild-type cells. In the cells of CHM1, only the larger band corresponding to REP2 (arrow) is detected. This was confirmed using an antibody specific to REP1, which detected a single band in wild-type cells (arrowhead) and no band in CHM1 cells. REP1, Rab escort protein 1.