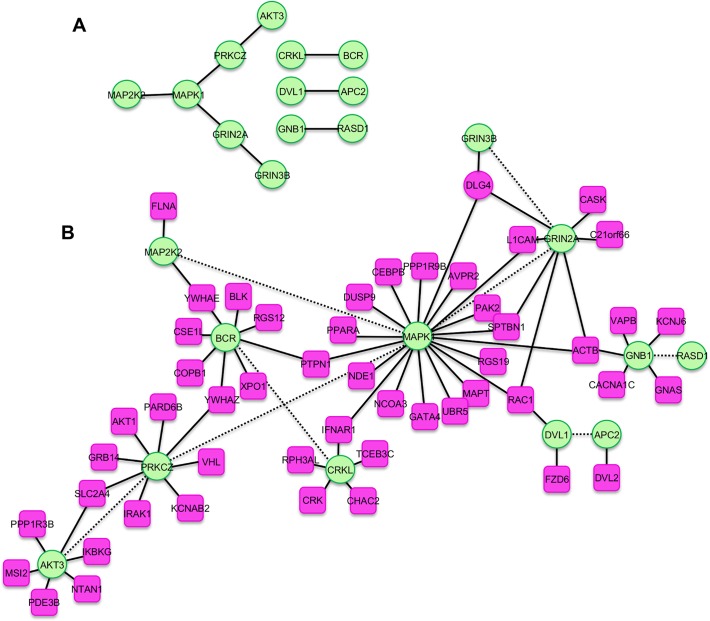
Fig 3. Molecular pathways serially identified among patients with microcephaly phenotypes in two large cohorts.
(A) 12 copy variant genes drawn from 14 of 27 Nijmegen patients with Microcephaly that were identified using multiple functional genomics methods (KEGG, Gene Expression and GO) and cluster strongly (p = 0.04) in the Dapple protein-protein interaction network. (B) Genes (n = 51; Red)) that were copy number variant in 30 of 71 Decipher patients with Microcephaly were found to possess a significant number of interactions with the genes from panel A (Green) (p = 0.04), forming an extensive and intertwined microcephaly-associated molecular network.