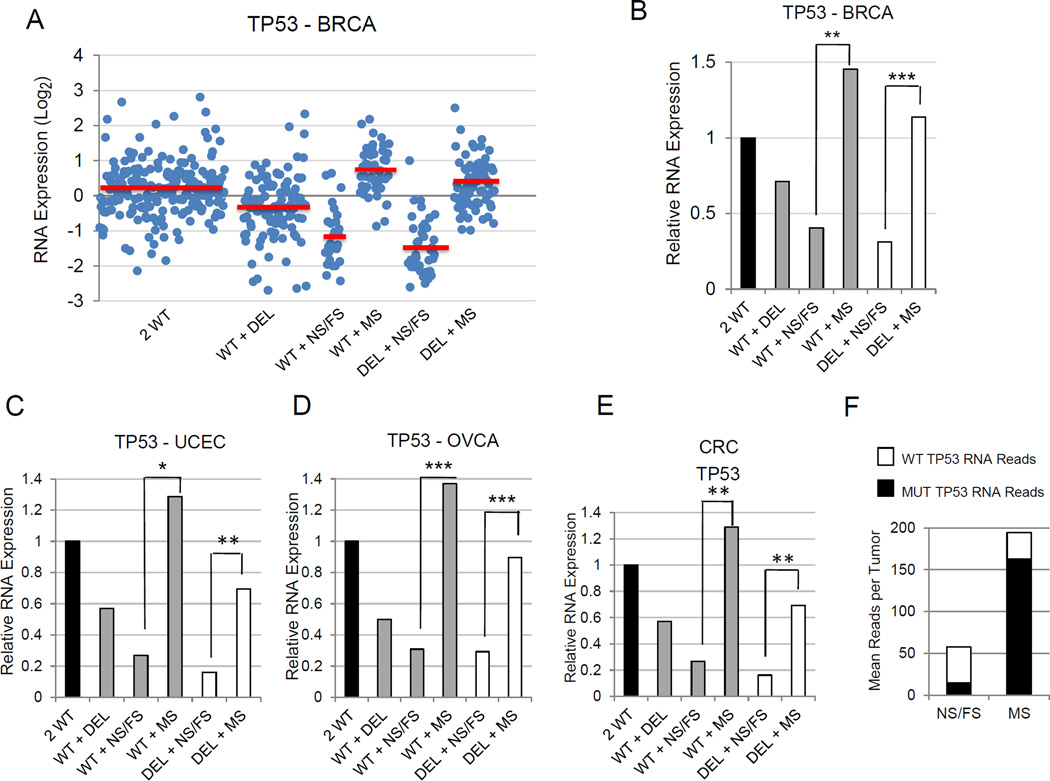
Figure 5.
TP53 RNA expression is significantly reduced in tumors with TP53 frameshift and nonsense mutations. (A) Scatter plot showing TP53 RNA expression in individual breast carcinomas sorted by TP53 allele status. Mean TP53expression values for each TP53 allele group are indicated by red bars. (B) Bar graph simplifying data from panel A showing TP53RNA expression levels in breast carcinomas segregated by TP53 allele status. Asterisks in panels B-E indicate the relative significance of the difference in expression of the designated TP53 categories. *P < 0.05; **P < 1E-05; ***P < 1E-25. (C–E) Bar graphs showing relative expression of TP53 RNA in endometrial carcinomas (C), ovarian serous adenocarcinomas (D), and colorectal carcinomas (E) by TP53 allele status. (F) Low levels of mutant TP53 RNA levels are found in tumors with nonsense or frameshift TP53 mutations, consistent with nonsense-mediated mRNA decay. Total mutant and wildtype TP53 RNAseq reads were analyzed in individual tumors with TP53 mutations from four different cancers (BRCA, OVCA, UCEC, CRC). The tumors were stratified by those with nonsense or frameshift TP53 mutations and those with missense TP53 mutations. Mean numbers of RNA reads per tumor and relative ratios of wildtype and mutant RNA allele fractions are indicated.