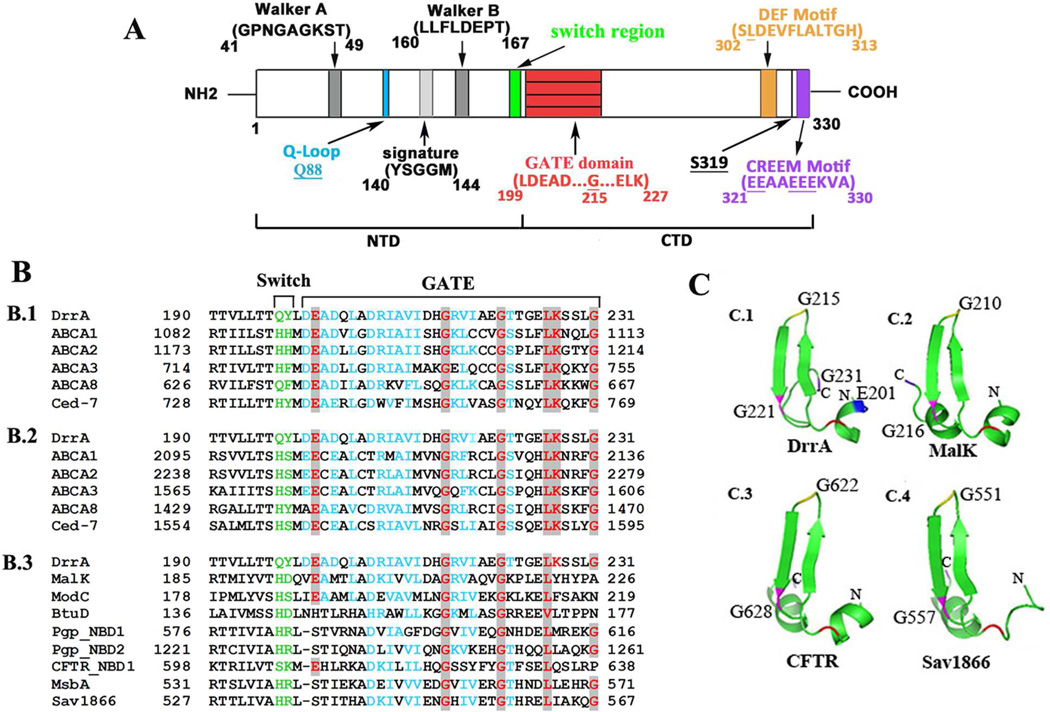
Fig. 1.
Sequence and structure analysis of the GATE domain. A, schematic representation of the conserved domains in DrrA. Numbers indicate the location of specific amino acid residues. B, amino acid alignment of GATE with homologs of the ABC superfamily. B.1 and B.2, alignment of GATE with the corresponding region of NBD 1 and 2 of close homologs of the DRA family. B.3, alignment of GATE with diverse ABC proteins. Switch motif, green; highly conversed residues, red; similar residues, blue. C, tertiary structure of GATE domain. C.1, structure of the GATE domain derived from an established model of DrrA [13]. Key residues E201, G215, G221, and G231 are marked in blue, yellow, pink and purple, respectively. Equivalent residues in the structures of ABC homologs (C.2, C.3 and C.4) have the same labels. The PDB accession numbers and source of each protein are provided in supplementary data.