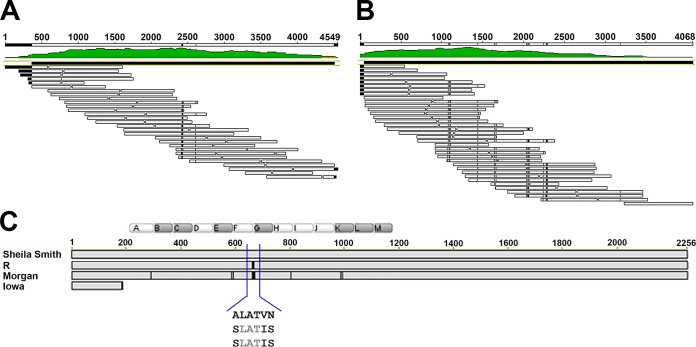
FIG 3.
Sequencing of the rOmpA repeat region and alignment of rOmpA. The transposon insertion kit EZ-Tn5 (DHFR-1) (Epicentre) was used to sequence and arrange the rOmpA series of repeats. This method generated randomly inserted primer binding sites that allowed for extended sequence reads and assembly of the repeats. (A and B) Sequence coverage from E. coli clones for the R (A) and Morgan (B) strains. Transposon insertion sites from which sequencing was primed are shown as gaps (H) in the sequence. (C) Sequence alignment of R and Morgan strains to Sheila Smith reveals seven shared SNPs, resulting in three nonsynonymous amino acid changes at the beginning of the type II G repeat. Clear boxed letters represent type I repeats, and shaded boxes represent type II. The area of shared changes is expanded to show hydrophobic A-to-hydrophilic S residue substitution. Morgan also contains 17 other SNPs, 9 of which result in 8 amino acid changes (two SNPs in one codon).