Fig 3. Expression analysis of sORF2.
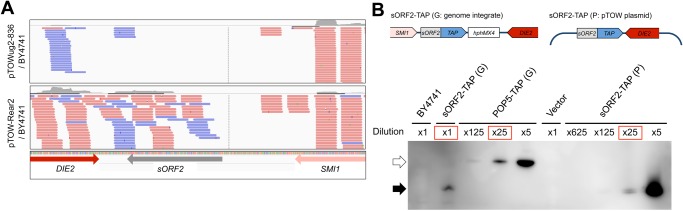
- RNAseq analysis of the sORF2 region of the strain BY4741 with the control vector (pTOWug2–836) and pTOW-Rear2. Parts of the detected reads are shown. The locations of DIE2, sORF2, and SMI1 are indicated.
- Western blot analysis of sORF2 using TAPtag. Expression of sORF2-TAP from the genomic region or plasmids was detected using peroxidase anti-peroxidase soluble complex. BY4714 is a negative control strain without any TAP-tagged protein expressed. Vector is another negative control, in which BY4741 harbors an empty vector (pTOWug2–836). Cells of BY4741, sORF2-TAP (genome), and POP5-TAP (genome) were cultivated in YPD medium; cells of Vector and sORF2-TAP (plasmid) were cultivated in SC—Ura medium. Dilution indicates the fold-dilution of the cellular lysate applied to the gel. Red-squared dilutions were used to calculate the expression levels of TAP-tagged proteins. The white arrowhead indicates the expected molecular weight of Pop5-TAP protein (39.6kDa), and the black arrowhead indicates the one of sORF2-TAP (27.1 kDa). Structures of sORF2-TAP constructs are shown.
