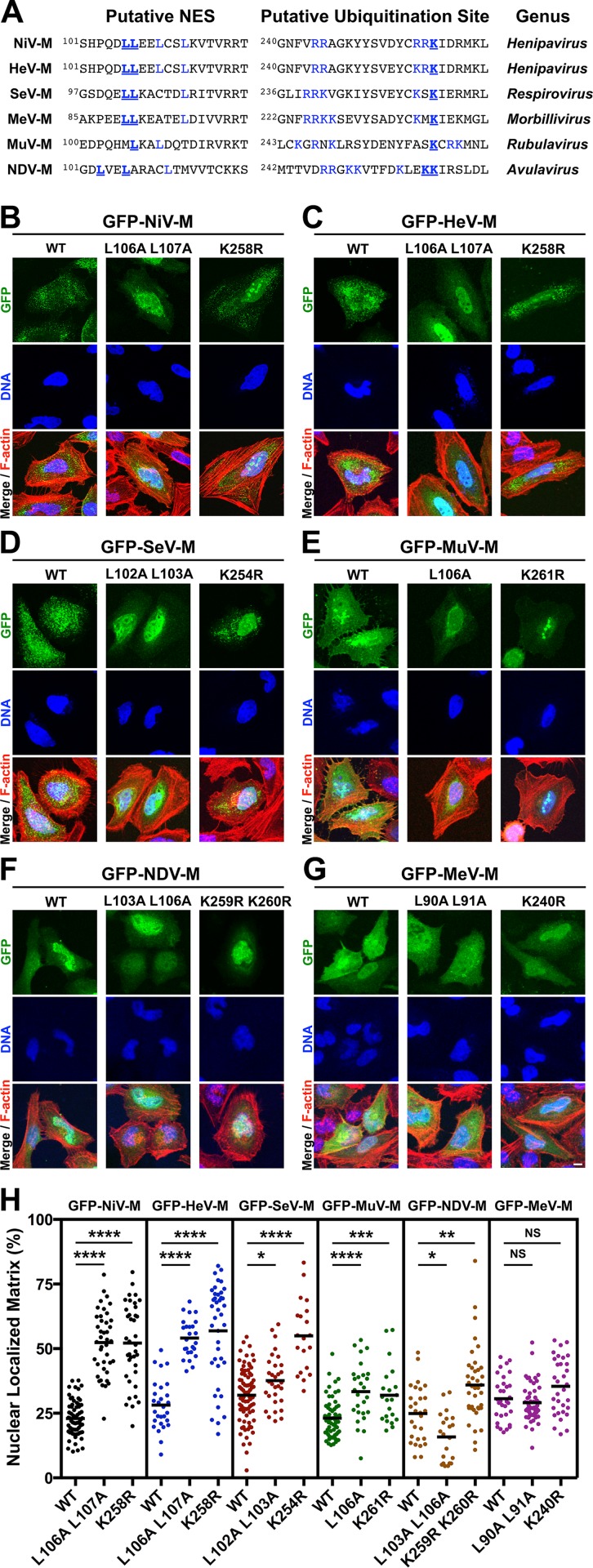
Fig 2. Mutational analysis of the role of a putative NES and a lysine within the NLSbp in nuclear export of Paramyxovirinae matrix proteins.
(A) Alignment of Paramyxovirinae M sequence motifs that correspond to NiV-M's leucine-rich NES and NLSbp, which contains a putative ubiquitinated lysine. Predicted critical residues are colored blue. Residues mutated in this study are also underlined in bold font. (B-G) Extended Focus (maximum intensity projection) views of 3D confocal micrographs of HeLa cells transfected with WT, or the indicated mutant GFP-tagged (B) NiV-M, (C) HeV-M, (D) SeV-M, (E) MuV-M, (F) NDV-M or (G) MeV-M. Cells were counterstained with DAPI to visualize nuclear DNA, blue, and fluorescent phalloidin to visualize the F-actin cytoskeleton, red. Scale bar 10 μm. (H) The amount of nuclear M fluorescence per cell was quantified from 3D reconstructed confocal micrographs. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001; NS, not significant by one-way ANOVA with Bonferroni adjustment for multiple comparisons.