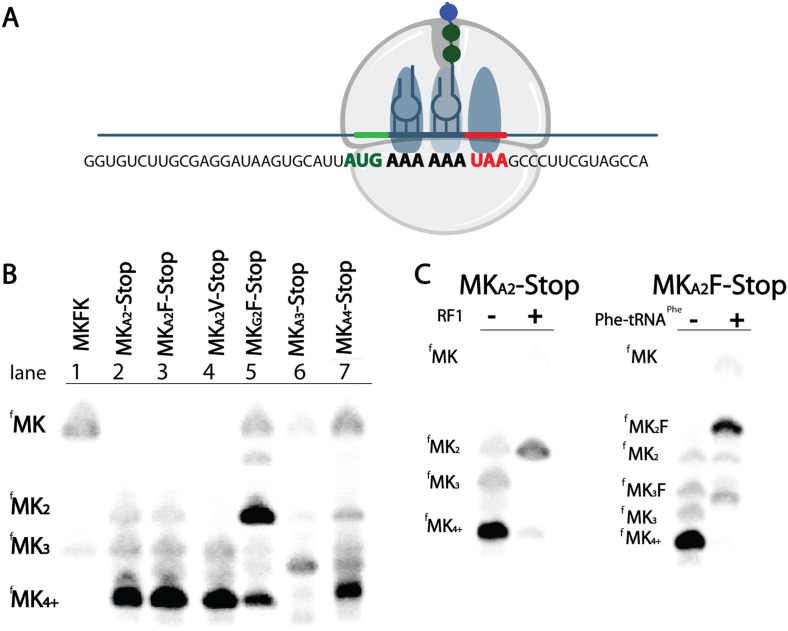
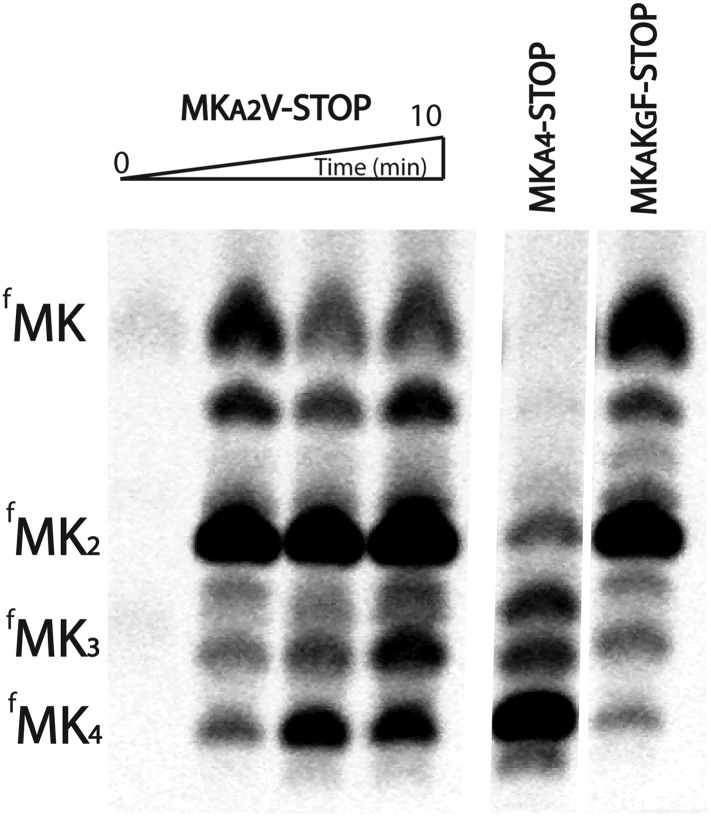
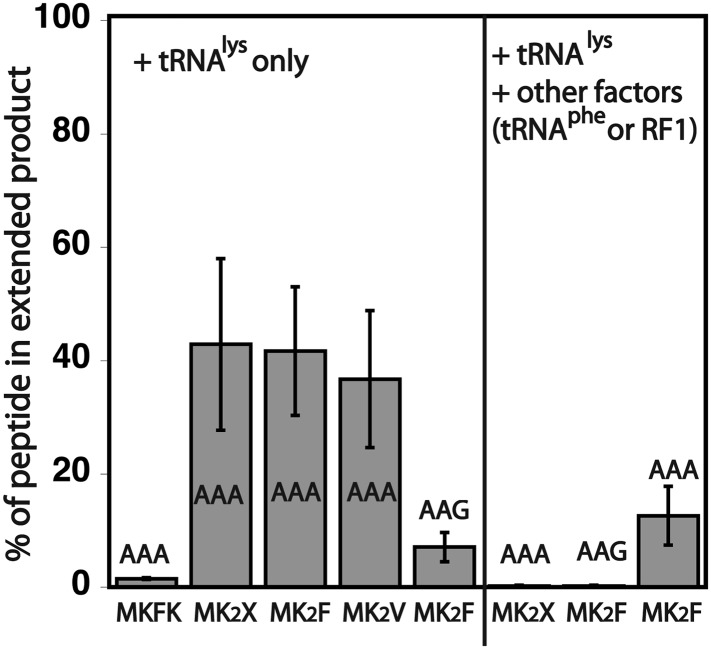
Figure 3. E. coli ribosomes add extra lysines on messages containing two sequential AAA, but not AAG, lysine codons.
(A) Illustration of the ribosome on the entire MKA2-Stop message. (B) eTLCs showing the peptide products resulting from translation of indicated messages with Lys-tRNAlys (but no other tRNAs or release factors) present. (C) eTLC displaying the peptide products resulting from the translation of indicated messages in the presence of Lys-tRNALys alone, or in the presence of Lys-tRNALys + factors (either RF1 or Phe-tRNAPhe) necessary for messages to be fully translated.