Significance
Flagella are bacterial organelles of locomotion. The number and location of flagella (flagellation pattern) are species specific and represent one of the earliest taxonomic criteria in microbiology. During each round of cell division, bacteria reproduce their flagellation pattern. FlhG is essential to a variety of flagellation patterns (e.g., polar, lateral) by yet-unknown mechanisms. We show that FlhG is an MinD-like ATPase that interacts with the flagellar C-ring proteins FliM/FliY in a nucleotide-independent manner. FlhG activates FliM/FliY to assemble with the C-ring protein FliG. FlhG-driven assembly of the FliM/FliY/FliG complex is strongly enhanced by ATP and lipids. We identify an underappreciated structural diversity of flagellar building blocks that contribute to formation of different flagellation patterns.
Keywords: flagellum, FlhG, C-ring, Bacillus, Shewanella
Abstract
The number and location of flagella, bacterial organelles of locomotion, are species specific and appear in regular patterns that represent one of the earliest taxonomic criteria in microbiology. However, the mechanisms that reproducibly establish these patterns during each round of cell division are poorly understood. FlhG (previously YlxH) is a major determinant for a variety of flagellation patterns. Here, we show that FlhG is a structural homolog of the ATPase MinD, which serves in cell-division site determination. Like MinD, FlhG forms homodimers that are dependent on ATP and lipids. It interacts with a complex of the flagellar C-ring proteins FliM and FliY (also FliN) in the Gram-positive, peritrichous-flagellated Bacillus subtilis and the Gram-negative, polar-flagellated Shewanella putrefaciens. FlhG interacts with FliM/FliY in a nucleotide-independent manner and activates FliM/FliY to assemble with the C-ring protein FliG in vitro. FlhG-driven assembly of the FliM/FliY/FliG complex is strongly enhanced by ATP and lipids. The protein shows a highly dynamic subcellular distribution between cytoplasm and flagellar basal bodies, suggesting that FlhG effects flagellar location and number during assembly of the C-ring. We describe the molecular evolution of a MinD-like ATPase into a flagellation pattern effector and suggest that the underappreciated structural diversity of the C-ring proteins might contribute to the formation of different flagellation patterns.
Most bacteria move by flagella. The flagellar architecture is conserved and can be divided into the cytoplasmic C-ring, the basal body, the rod, and the exterior hook and filament structures (1). Bacterial species differ in the number and arrangement of their flagella (flagellation pattern) (2). However, the mechanisms that allow bacteria to establish their specific flagellation patterns reproducibly during each cell division are poorly understood. The protein FlhG (also known as “YlxH,” “MinD2,” “FleN,” or “MotR”) is essential for the correct flagellation pattern of polar- (3–5), lophotrichous- (6), amphitrichous- (7), and peritrichous-flagellated bacteria (8, 9). Deletion of flhG in polar-flagellated bacteria leads to hyperflagellation and impaired motility (3–5). In the amphitrichous-flagellated Campylobacter jejuni, ∼40% of the cells of a ΔflhG strain exhibited more than one flagellum at one pole and were impaired in motility (7). The peritrichous-flagellated bacterium Bacillus subtilis exhibits ∼26 flagellar basal bodies arranged symmetrically around midcell in a gridlike pattern (8). Furthermore, flagella are discouraged at the cell pole. Deletion of flhG does not result in swimming or swarming defects, although multiple flagella appear in tufts from constrained loci on the cell, and flagellar basal bodies often are aggregated (8). FlhG acts in concert with the signal recognition particle (SRP)-GTPase FlhF (10–14) that recruits the flagellar protein FliF to the cell pole in the polar-flagellated Vibrio cholerae (15). FlhG is predicted to belong to the MinD/ParA ATPase family (6, 16) whose characterized members act in orchestrated spatiotemporal processes (e.g., cell-division site determination and plasmid/chromosome partitioning (summarized in ref. 17). Together with MinC, MinD constitutes the conserved center of the Min system which regulates bacterial cell division by restricting cytokinetic Z-ring assembly to midcell (reviewed in ref. 18). MinD forms ATP-dependent homodimers (19) that interact with the inner membrane through a C-terminal amphipathic helix (membrane-targeting sequence, MTS) (20, 21). By this mechanism, MinD recruits MinC to the membrane where MinC inhibits polymerization of FtsZ into the Z-ring (22). Interestingly, Campylobacter jejuni does not contain a Min system, and FlhG is involved in flagellation pattern control and regulation of cell division (7). In contrast to MinD, the molecular framework in which the putative MinD-like ATPase FlhG controls flagellation is unknown. Also, it is enigmatic how conserved homologs of FlhG can control different flagellation patterns in different species. Here, we investigated the mechanism and function of FlhG in the Gram-positive, peritrichous-flagellated B. subtilis (Bs) and the Gram-negative, polar-flagellated Shewanella putrefaciens (Sp).
Results
FlhG Is a MinD-Like ATPase.
First, we determined the crystal structure of FlhG. We used FlhG from the moderate thermophile Geobacillus thermodenitrificans (Gt), because thermophilic proteins tend to crystallize better than their mesophilic counterparts (Fig. S1A). The crystal structure of GtFlhG was determined at 2.8-Å resolution (Table S1) and lacked residues 1–20 and 265–274, likely because of flexibility. The structures of GtFlhG and Escherichia coli (Ec) MinD [Protein Data Bank (PDB) ID code 3Q9L] superimpose with an rmsd of 2.3 Å2 for 160 Cα atoms, revealing a conserved protein core with minor deviations in the helical periphery (Fig. 1A and Fig. S1B). Structural differences between GtFlhG and EcMinD are most pronounced in an extension of helix α7 and a loop that replaces a helical turn between α6 and β7 found in MinD. Our structural analysis shows conservation of the key active-site motifs required for ATP–Mg2+ binding and hydrolysis in FlhG and MinD (Fig. S1C). HPLC-based ATP hydrolysis assays proved that GtFlhG is an ATPase with an activity of 51.2 ± 2.4 nmol (ATP)⋅h−1⋅nmol(enzyme)−1 under our in vitro assay conditions. A GtFlhG D60A variant, which disrupts a catalytically relevant magnesium-binding site, lacks catalytic activity (Fig. 1B) (19, 23). Thus, we demonstrate that FlhG is a MinD-like ATPase.
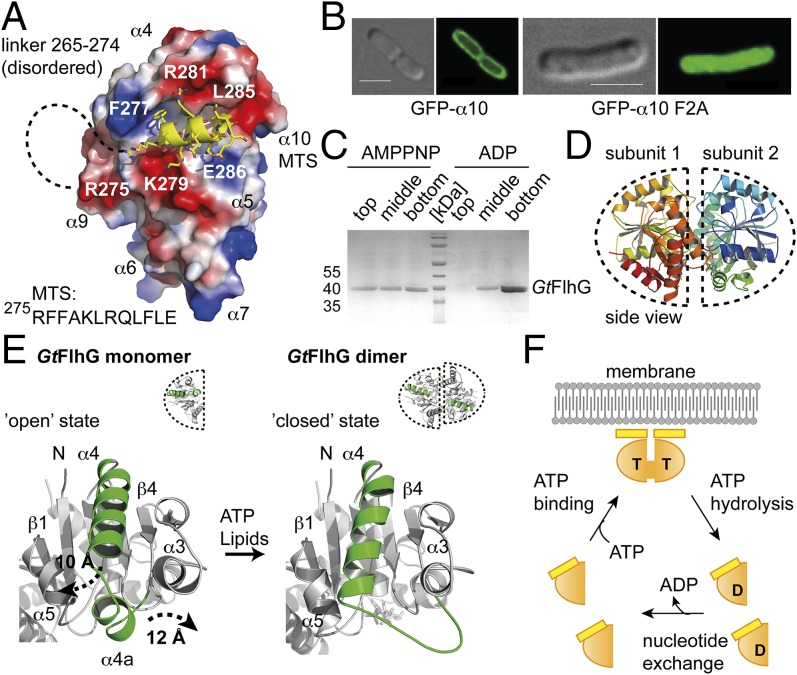
Fig. 1.
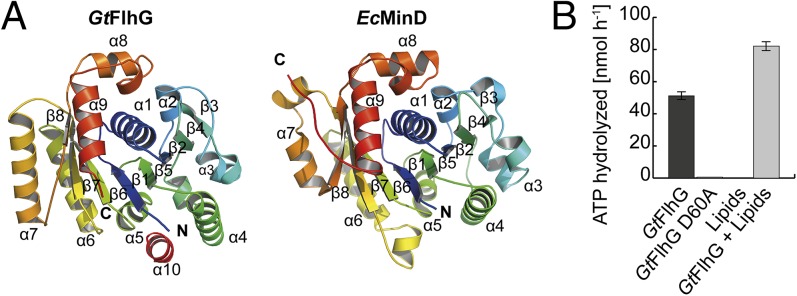
Crystal structure of the MinD ATPase FlhG. (A) Cartoon representation of the crystal structures of GtFlhG (this study, Left) and EcMinD (PDB ID: 3Q9L, Right). Both structures are rainbow-colored from the N to the C terminus as indicated by “N” and “C,” respectively. (B) ATPase activity of GtFlhG and the GtFlhG D60A variant (in nanomoles per hour) in the absence or presence of lipids. GtFlhG (20 µM) was incubated with 2 mM ATP at 37 °C for 1 h.
Lipid Interaction of FlhG is ATP Dependent and Mediated by its C-Terminal MTS.
The interaction of MinD with membrane lipids is mediated by its MTS (20, 21). Its amphipathic nature is conserved between FlhG and MinD proteins (Fig. S1C). The GtFlhG structure resolves the putative MTS (helix α10) that packs into a hydrophobic groove formed by helices α4 and α5 of FlhG (Fig. 2A). To investigate whether α10 also serves as an MTS in FlhG, we fused α10 of GtFlhG to the C terminus of GFP (GFP-α10) and investigated its subcellular localization. GFP-α10 localized predominantly at the plasma membrane (Fig. 2B, Left). Substitution of the conserved phenylalanines 276 and 277 in α10 by alanines (GFP-α10-F2A) abolished its membrane association (Fig. 2B, Right). Thus, helix α10 of FlhG constitutes an MTS. To validate these findings biochemically, we performed flotation assays of GtFlhG with large unilamellar vesicles (LUVs) composed of 70% phosphatidylethanolamine (PE) and 30% phosphatidylglycerol (PG) as a model membrane (24). In short, the protein was incubated with LUVs and applied to gradient ultracentrifugation. Upon binding to LUVs, FlhG relocates to the top of the gradient, and unbound protein remains at the bottom of the gradient. In addition, we investigated the influence of ADP and ATP [mimicked by the nonhydrolysable ATP-analog 5′-adenylyl-imidodiphosphate (AMPPNP)] on the lipid binding of FlhG. Although GtFlhG interacted with LUVs in the presence of AMPPNP, no interaction was observed with ADP (Fig. 2C). These results demonstrate that FlhG interacts with lipids and imply that ATP-binding to FlhG is required for its lipid interaction.
Fig. 2.
Lipid- and ATP-dependent homodimerization of FlhG. (A) Electrostatic surface view of GtFlhG with the MTS shown in yellow. Dashed lines indicate the disordered linker (residues 265–274) connecting ATPase and the MTS. (B) In vivo fluorescence micrographs of GFP-α10 and GFP-α10-F2A show that α10 of GtFlhG is a functional MTS. (Scale bars, 2 µm.) (C) Coomassie-stained SDS/PAGE of the flotation assay of GtFlhG with LUVs in the presence of ADP and AMPPNP (from top to bottom fractions). Note: GtFlhG interacting with LUVs is found in the top fraction. (D) Cartoon representation of the GtFlhG homodimer. Dashed lines indicate each monomer. Note: Although ATP was added before crystallization, the crystal structure of the GtFlhG homodimer has only ADP bound in its active sites, likely because of residual ATPase activity during crystal growth (3–4 wk). (E) Structural differences between the monomeric and dimeric states of GtFlhG. Major conformational changes are shown in green. (F) Model of the FlhG ATPase mechanism (orange) showing the ATP (T)-dependent homodimerization and expulsion of the MTS (yellow), membrane interaction of the homodimer through the MTS, and ATP hydrolysis-dependent dissociation of the homodimer.
Crystal Structure of the FlhG Homodimer.
By analogy to MinD (19), we reasoned that FlhG also might form ATP-dependent homodimers. To increase the success of crystallizing the FlhG homodimer, we used a catalytically inactive GtFlhG variant (i.e., D60A; Fig. 1B) that was used previously to crystallize the MinD homodimer (19). The crystal structure of GtFlhG-D60A was determined at 1.9-Å resolution (Fig. 2D and Table S1). The GtFlhG and EcMinD homodimers resemble an ellipselike shape with similar dimensions of 60 Å, 45 Å, and 40 Å and significant structural homology (rmsd of 2.8 Å2 over 329 Cα atoms; Fig. 2D and Fig. S2 A and B). The subunits are arranged in the same face-to-face orientation, and no differences in the active sites exist (for a detailed structure comparison, see Fig. S2 C–E). Electron density corresponding to the MTS was clearly visible in the GtFlhG monomer but was lacking for the MTS in the homodimer. To understand this phenomenon better, we compared the structures of the GtFlhG monomer and homodimer (Fig. 2E). In particular, helix α4 and the preceding helical segment α4a undergo significant structural rearrangements of 10 Å and 12 Å, respectively. Also, α4a looses its helical propensity in the homodimer and appears as an elongated loop that contributes to the dimer interface. However, helix α4 (together with α5) also establishes the hydrophobic groove that harbors the MTS in the monomeric state. Our structural comparison shows that the conformational state of helix α4 in the homodimer closes the MTS-binding groove and therefore fosters solvent and lipid accessibility to the MTS. Conversely, binding of the MTS to the hydrophobic groove precludes the movement of α4 and α4a into a configuration suitable for homodimer formation. Thus, MTS-mediated lipid binding and ATP-dependent homodimerization of FlhG are highly dependent on each other. We conclude that FlhG can switch between two mutually exclusive states: (i) an ATP-bound homodimer that associates with the plasma membrane through its MTS and (ii) an ADP (or nucleotide-free) monomer that is unable to interact with the membrane (Fig. 2F).
Nucleotide-Independent Interaction of FlhG with the C-Ring Proteins FliM and FliY.
To understand the function of BsFlhG, we performed pulldown assays in whole-cell lysates of B. subtilis using purified GST-tagged BsFlhG. Mass spectrometry suggested the proteins FliM and FliY as binding partners of FlhG (Fig. S3A). FliM interacts directly with FliY (a homolog of FliN), and, together with FliG, all three constitute the flagellar C-ring (reviewed in ref. 1). To validate these findings, we performed in vitro pulldown assays using GST-FlhG and the FliM/FliY complex that were overexpressed in E. coli and purified by Ni-ion affinity and size-exclusion chromatography (SEC). The interaction of FlhG and FliM/FliY was validated in vitro for the proteins from B. subtilis and G. thermodenitrificans (Fig. 3A and Fig. S3 B and C). Finally, we reconstituted the GtFlhG/FliM/FliY complex on SEC (Fig. S3D). Proteins from G. thermodenitrificans showed more stable biochemical behavior. To correlate biochemical behavior with the structural data, we decided to perform our in vitro experiments with GtFlhG and GtFliM/FliY. Nucleotides (i.e., ADP, ATP, or AMPPNP) did not affect the interaction of GST-GtFlhG with GtFliM/FliY (Fig. 3A and Fig. S3E). Furthermore, two GtFlhG variants, which are defective in ATP and magnesium binding (i.e., K36Q and D60A, respectively), retained their ability to bind GtFliM/FliY (Fig. S3F). Thus, we show that FlhG interacts with a complex of the flagellar C-ring proteins FliM and FliY in a nucleotide-independent manner. To specify which part of the FliM/FliY complex provides the binding site for FlhG, we used hydrogen-deuterium exchange (HDX) mass spectrometry. This method allows rapid determination of protein–protein interfaces (24). Specifically, GtFliM/FliY was incubated with and without GtFlhG and, after completion of the HX-labeling reaction, was digested with pepsin. Peptic peptides were analyzed by electrospray ionization-mass spectrometry. HDX mass spectrometry suggested that FlhG binds to the N terminus of FliY (i.e., amino acids 12–28 of GtFliY) (Fig. 3B and Fig. S4). Indeed, a GtFliM/FliY complex in which FliY lacked its N terminus (FliY-ΔNtr), was unable to interact with GtFlhG (Fig. 3C). FliY-Ntr contains a highly conserved amino acid motif (i.e., the EIDAL motif), which also is present in the N-terminal region of FliM (FliM-Ntr) (25). However, the absence of the FliM-Ntr at the GtFliM/FliY complex did not influence its ability to bind to GtFlhG (Fig. 3C). Thus, FliY-Ntr primarily mediates the interaction of FlhG with the FliM/FliY complex.
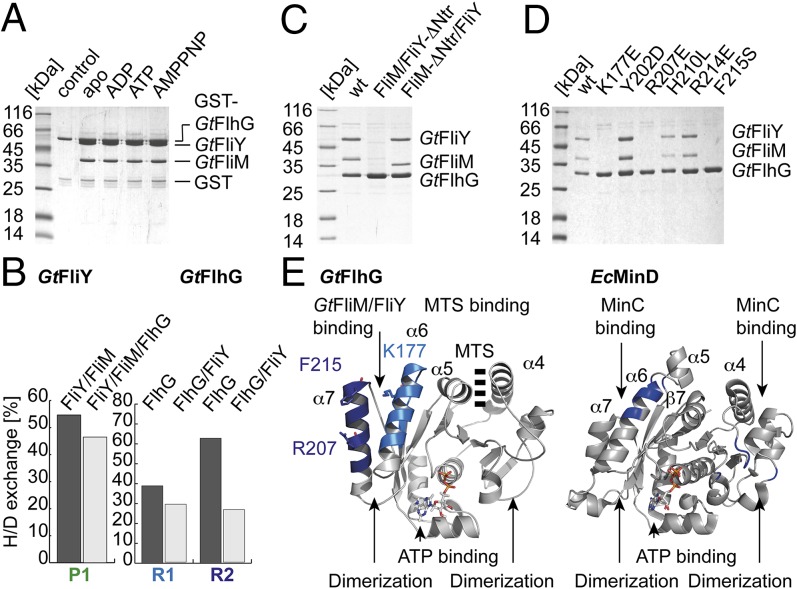
Fig. 3.
FlhG interacts with the flagellar C-ring proteins FliM/FliY. (A) Coomassie-stained SDS/PAGE of an in vitro pulldown assay of GST-GtFlhG and GtFliM/FliY in the absence and presence of ADP, ATP, or AMPPNP. (B) Deuterium incorporation of depicted peptides of free protein and the dimeric GtFlhG/FliY complex are given in percent H/D exchange. Decreased deuterium content upon complex formation indicates potential interfaces (peptides GtFliY R1: DALLRGMDDSDHVPALH; GtFlhG P1: TDAYAMMKYMHAAGSEAPFSV and P2: VFERLKHVTGRFLNKD). (C) Coomassie-stained SDS/PAGE of an in vitro pulldown assay using (His)6-tagged GtFlhG, GtFliM/FliY, and GtFliM/FliY variants lacking the FliY-Ntr and FliM-Ntr. (D) Coomassie-stained SDS/PAGE of an in vitro pulldown assay using different (His)6-tagged GtFlhG variants and the GtFliM/GtFliY complex. (E) Major differences that allow FlhG (Left) and MinD (Right) to bind FliM/FliY and MinC, respectively, are shown in blue.
Molecular Evolution of a MinD-Like ATPase into a Flagellar Biogenesis Factor.
Next, we wanted to determine which part of FlhG interacts with FliM/FliY. HDX experiments identified two regions in GtFlhG (Fig. 3B and Fig. S5) that are located at helices α6 and α7 (Fig. 3E). To verify this observation, we varied solvent-exposed amino acid residues at helices α6 and α7 in GtFlhG and probed their binding to GtFliM/FliY by pulldown assays (Fig. 3D). Three residues (Lys177, Arg207, and Phe215), which are located on helices α6 and α7 of FlhG, are essential for FliM/FliY binding (Fig. 3 D and E). As described above, this region represents the major difference between FlhG and MinD. In MinD, the corresponding region is essential for MinC binding (19). Accordingly, GtFlhG is unable to interact with GtMinC (Fig. S3G). Therefore, the described structural differences between MinD and FlhG are the basis for their different interactions during cell division (via MinC) and flagella assembly (via FliM/FliY), respectively (Fig. 3E). These differences also illustrate how evolution relies on the modification of existing structures for new functions.
Subcellular Behavior of FlhG in B. subtilis.
To place our findings in a biological context, a BsFlhG-YFP fusion protein was introduced into the native genomic locus in B. subtilis. BsFlhG-YFP was monitored using high-resolution fluorescence microscopy in cells that were grown to the exponential or stationary phase. The majority of BsFlhG-YFP localized at the membrane in distinct foci (Fig. 4A). These foci could be divided into two main subpopulations that either were stationary for more than 5 s or were dynamic in the millisecond range (n = 200) (Fig. 4A and Movie S1). Stationary foci did not show any change in location with an average resting time of 29.0 ± 22.6 s (n = 61). The ratio of stationary to dynamic foci was 23.4:76.6%, indicating that the majority of FlhG is highly mobile. To investigate the cellular function of FlhG further, we used a strain carrying BsFlhG-YFP and the flagellar C-ring protein FliM C terminally fused to CFP (FliM-CFP). As described earlier (8), FliM-CFP is functional and is almost completely static at the membrane, indicating its incorporation into the flagellum. We observed colocalization of FliM and a subfraction of FlhG that occurred for ∼30 s (average colocalization: 33.0 ± 20.1 s; n = 31) (Fig. 4 B and C and Movies S2, S3, and S4). These data corroborate our biochemical analysis and indicate that the static fraction of FlhG might be involved in allocating flagellar C-ring proteins in B. subtilis.
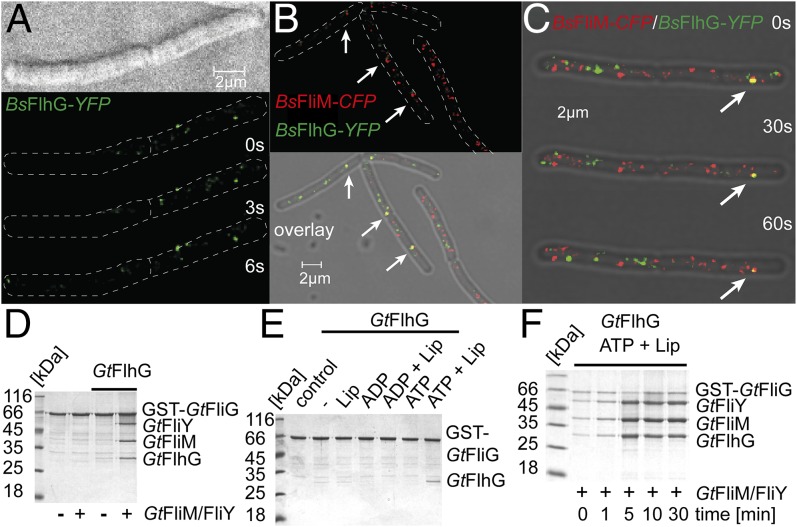
Fig. 4.
Physiological role of FlhG. (A) Localization of a BsFlhG-YFP fusion protein in B. subtilis displays distinct foci at the membrane. (B and C) Localization of FlhG and FliM was detected in B. subtilis carrying BsFlhG-YFP (green) and BsFliM-CFP (red). (D) Coomassie-stained SDS/PAGE of an in vitro pulldown assay of GST-GtFliG with GtFliM/FliY in the absence and presence of GtFlhG. (E) Coomassie-stained SDS/PAGE investigating the ability of GtFlhG to bind to GST-GtFliG and its dependence on lipids, ADP, ADP+lipids, ATP, and ATP+lipids. (F) Coomassie-stained SDS/PAGE of a time-resolved pulldown assay (0, 1, 5, 10, and 30 min) investigating the binding of GtFliM/FliY and GtFlhG to GST-GtFliG in presence of ATP and lipids.
The FlhG ATPase Enhances Formation of the FliM/FliY/FliG Complex.
The FliM/FliY complex interacts with FliG within the C-ring of a mature flagellum. Therefore, we performed in vitro pulldown assays using GST-tagged GtFliG (GST-GtFliG), GtFliM/FliY, and GtFlhG that were produced in E. coli and purified by Ni-ion affinity and SEC. First, we analyzed the in vitro binding of purified GtFliM/FliY to GST-GtFliG in the absence or presence of GtFlhG. In the absence of FlhG, FliM/FliY did not interact with GST-FliG. However, the presence of FlhG yielded an almost stoichiometric binding of FliM/FliY and FlhG to GST-FliG (Fig. 4D). These data show that FlhG mediates efficient formation of the FliM/FliY/FliG complex in vitro. Delivery of FliM/FliY to FliG would position FlhG in close proximity to the membrane, where FlhG could interact with membrane lipids through its MTS and dimerize in an ATP-dependent manner. Possible spatial restraints of FlhG by the membrane on the one hand and by FliM/FliY on the other suggested that FlhG also might contact FliG. Therefore, we investigated whether FlhG could interact with FliG and whether this interaction would be nucleotide and/or lipid dependent. GST-GtFliG and GtFlhG were incubated in the absence and presence of ADP, ATP, lipids, and their combinations. FliG/FlhG interaction was observed in the presence of ATP+lipids, but no interaction occurred with lipids alone, ADP, ADP+lipids, or ATP (Fig. 4E). Thus, we conclude that lipid-mediated and ATP-dependent homodimerization of FlhG is a prerequisite for its interaction with FliG. These findings also indicate that ATP and lipids might influence the FlhG-mediated assembly of FliM/FliY into FliG. Therefore, we analyzed the GtFlhG-mediated binding of GtFliM/FliY to GST-GtFliG at different time points in the presence of ATP and lipids (Fig. 4F and Fig. S3H). We observed a gradual increase of FliM/FliY/FlhG binding to GST-FliG over time. After ∼5 min, we observed a stoichiometric ratio exceeding 1 of FliM/FliY/FlhG compared with GST-FliG, suggesting the oligomeric assembly of FliM/FliY structures at GST-FliG. These findings strongly indicate that FlhG coordinates the assembly of FliM/FliY to FliG in an ATP- and lipid-dependent manner.
FlhG Is Essential for the Polar Flagellation Pattern in S. putrefaciens.
FlhG also is conserved among other bacteria with polar, lophotrichous, and amphitrichous flagellation (2, 11, 26). To understand whether the functional role and mechanistic principles of FlhG could be applied to other flagellated bacteria, we chose the Gram-negative γ-proteobacterium S. putrefaciens (Sp) CN-32. This species possesses gene clusters encoding two distinct flagellar systems (27, 28). Expression of the primary cluster, which also comprises flhG, leads to production of a single flagellar filament at the cell pole (Fig. 5A, Upper). Subpopulations expressing the secondary system develop a secondary flagellum at a lateral position (27). An in-frame deletion of flhG in S. putrefaciens resulted in cells that were hardly motile and were hyperflagellated (2–16 filaments) at the cell pole (Fig. 5A, Lower and Fig. S6 A and B). Western blot analysis confirmed successful gene deletion (Fig. S6C). This observation agrees with the hyperflagellation phenotypes of flhG deletions in other polar-flagellated γ-proteobacteria (12, 15, 26, 29). Using fluorescence labeling on FlgE2-Cys (the T242C mutant) as a marker for the secondary lateral flagellum (30, 31), we found that the size, number, and position of the secondary flagellum did not change significantly in ΔflhG mutants (14.1% wild-type; 15.2% ΔflhG) (Fig. S6 D and E). Thus, we conclude that SpFlhG addresses the primary polar flagellum. SpFlhG exhibited ATPase activity (Fig. 5B), and strains bearing a hydrolysis-deficient FlhG variant (i.e., K29A) displayed a hyperflagellation phenotype comparable to that of the ΔflhG strain (Fig. S6A). Therefore, ATPase activity of SpFlhG is essential to restrict the number of polar flagella to one. Similarly, mutants within the C-terminal MTS of SpFlhG (i.e., with phenylalanines 275/276 replaced by alanines) exhibited a hyperflagellated phenotype (Fig. S6A). The presence of these proteins was verified by Western blot analysis (Fig. S6C). Thus, we confirmed the hallmark features of BsFlhG in Shewanella.
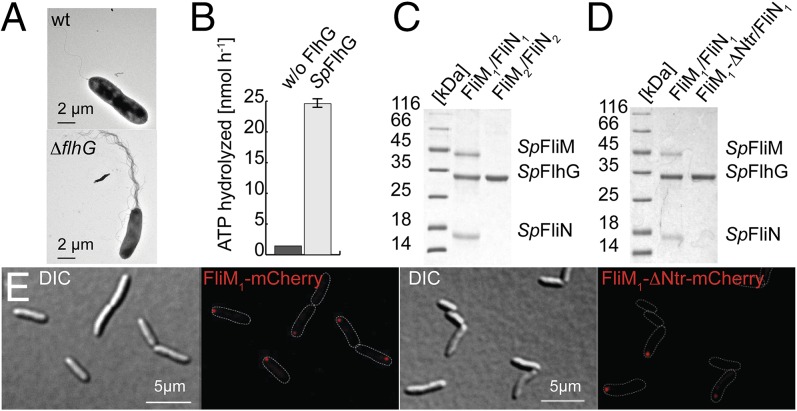
Fig. 5.
Role of FlhG in the polar- and lateral-flagellated S. putrefaciens. (A) Electron micrographs of S. putrefaciens (Upper) and its ΔflhG mutant (Lower). (B) SpFlhG ATPase activity (in nanomoles per hour). FlhG (100 µM) was incubated with 1 mM ATP at 37 °C for 30 min. (C) Coomassie-stained SDS/PAGE of an in vitro pulldown assay shows that (His)6-tagged SpFlhG interacts with SpFliM1/FliN1 but not with SpFliM2/FliN2. (D) Coomassie-stained SDS/PAGE of an in vitro pulldown assay shows that (His)6-tagged SpFlhG does not interact with an SpFliM1/FliN1 variant lacking the N-terminal 27 amino acids of FliM1 (FliM1-ΔNtr). (E, Left) FliM1-mCherry localizes in distinct foci at the cell pole of S. putrefaciens in 41% of the cells (n = 576). (Right) FliM1-ΔNtr-mCherry displays decreased polar localization (22% of cells; n = 456).
FlhG Interacts with the C-Ring Proteins FliM1/FliN1 of the Polar Flagellum in S. putrefaciens.
We reasoned that SpFlhG interacts with the C-ring proteins FliM1 and FliN1 (a homolog of FliY) of the polar flagellum but not with FliM2 and FliN2 of the lateral flagellum. In fact, our in vitro pulldown assays show that SpFlhG interacts with the FliM1/FliN1 complex but does not interact with FliM2/FliN2 (Fig. 5C). In S. putrefaciens, only FliM1, but not FliM2, harbors the conserved EIDAL motif within its N-terminal region (28). A FliM1/FliN1 complex that lacked the FliM1-Ntr did not interact with SpFlhG in vitro (Fig. 5D). Complementary in vivo experiments demonstrated that deletion of fliM1 in S. putrefaciens or removal of the FliM1-Ntr from FliM1 phenocopied an flhG deletion strain with respect to hyperflagellation and motility (Fig. S6 A and F). An S. putrefaciens strain carrying the red fluorescent reporter protein mCherry fused to the C terminus of FliM1 was constructed to determine its cellular localization. Distinct foci were observed at one pole in 41% of the cells (Fig. 5E, Left). Deletion of the FliM1-Ntr in the mCherry fusion protein reduces the proportion of cells exhibiting a correct localization to 22% (Fig. 5E, Right). Western blot analysis confirmed the expression of the fluorescently labeled proteins (Fig. S6G). Thus, we show that FlhG interacts with FliM1/FliN1 through a conserved motif at the N terminus of FliM1.
Discussion
Similarities and Differences Between MinD and FlhG.
FlhG preserved the hallmarks of MinD such as the overall fold, active site architecture, and ATPase activity. Both proteins form ATP-dependent homodimers that interact with membranes through a conserved MTS (this study and refs. 19–21). Therefore, like MinD, FlhG can cycle between two distinct states: a membrane-associated, ATP-bound homodimer and an ADP-bound (or nucleotide-free) monomer. In the E. coli Min system, MinE stimulates the activity of the MinD ATPase (32–34) and therefore releases MinD from the membrane. By analogy to MinD, we speculate that accessory factors influence the FlhG ATPase. FlhG deviates from MinD mainly at helices α6 and α7, which have been shown to mediate the interaction of MinD and MinC. The molecular differences at α6 and α7 create a new interface that enables FlhG to interact with the flagellar C-ring proteins but also prevent FlhG from binding MinC. Taken together, these observations provide the molecular basis for the evolution of a MinD-ATPase into a flagellar assembly factor.
Physiological Role of FlhG.
In B. subtilis, FlhG shows a highly dynamic behavior, suggesting that FlhG cycles between the cytoplasm and basal bodies/membrane. FlhG interacts with the FliM/FliY complex in a nucleotide-independent manner. Therefore, we speculated that FlhG might capture FliM/FliY in the cytoplasm and deliver the complex to FliG at nascent flagellar structures. Indeed, binding of FlhG to FliM/FliY seems to be a prerequisite for their productive interaction with FliG under our in vitro assay conditions. This observation suggests that FlhG induces a conformational change in FliM/FliY enabling their interaction with FliG. Whether FlhG-mediated assembly of FliM/FliY/FliG takes place in the cytoplasm before their attachment to FliF or whether FliG already resides at the nascent flagellar structure cannot yet be resolved. Furthermore, our in vitro assays show an FlhG-dependent assembly of oligomeric FliM/FliY structures at FliG in the presence of lipids and ATP that also are required for FlhG homodimerization. This observation suggests the homodimer has a role in coordinating the FliM/FliY/FliG assembly. However, the precise mechanism requires further research. A puzzling observation is that FlhG is not released from C-rings in vitro. In contrast, in vivo FlhG resides at the nascent flagellar structure/plasma membrane only temporarily, suggesting accessory factors are needed to release FlhG from the C-ring. In the peritrichous-flagellated B. subtilis, basal bodies appear in a gridlike pattern as monitored by a fluorescently labeled FliM (8). Deletion of flhG leads to severe aggregation of basal bodies. Deletion of flhG in S. putrefaciens, which contains a polar and lateral flagellar system, leads to an increased number of polar flagella, but neither the position of the lateral flagellum nor its number is changed. This finding is in agreement with other polar-flagellated bacteria in which deletion of flhG also increases the number of polar flagella (3–5). In S. putrefaciens, FlhG interacts with FliM1/FliN1 of the polar flagellar system but not with FliM2/FliN2 of the lateral one. Moreover, deletion of the FliM1-Ntr, which binds FlhG, results in an increased number of polar flagella in S. putrefaciens (as observed for ΔflhG). Also, in the amphitrichous-flagellated C. jejuni, a functional connection between FlhG and flagellar C-ring proteins was shown, although no direct protein–protein interaction data are available thus far (7). Taken together, these findings suggest that FlhG executes its role in the formation of the flagellation pattern during the assembly of the flagellar C-ring. However, at this point we cannot say whether FlhG delivers C-ring proteins to the nascent flagellum or if FlhG binding to these proteins blocks the assembly of a nascent flagellum. Both models are equally plausible, and further experiments are needed for clarification.
FlhG and the Diversity of Flagellar C-Ring Proteins.
FlhG interacts with FlhF in polar-, amphitrichous-, and peritrichous-flagellated bacteria (11, 12, 14), and a conserved motif at the N terminus of FlhG stimulates the GTPase activity of FlhF in B. subtilis (11). Although the FlhF–FlhG interaction seems to be conserved, subtle differences might contribute to the regulation of different flagellation patterns. Moreover, FlhG also might be involved in the regulation of flagellar gene expression. In Pseudomonas aeruginosa and V. cholerae, FlhG interacts with FleQ (also FlrA), a master regulator of flagella gene expression (35–37), adding another layer of complexity to the role of FlhG. However, C. jejuni and B. subtilis lack FleQ, and whether FlhG contributes to transcriptional regulation in these bacteria is unknown. In B. subtilis and S. putrefaciens FlhG binds to a region at the N termini of FliY and FliM1, respectively, that contains the conserved EIDAL motif. In mature flagella, this motif binds phosphorylated CheY that switches the flagellar rotor in response to chemosensory signals (Fig. S7) (38–40). Thus, FlhG might shield the CheY-binding site to prevent futile communication of the chemosensory system with nascent flagella. Further differences in the C-ring protein architectures exist (Fig. 6A): In S. putrefaciens, FliN1 and FliN2 constitute a conserved dimerization domain. However, FliM1 differs from FliM2 in the N-terminal extension to which FlhG binds (Fig. 6B). Thus, FlhG affects the polar flagellum but not the lateral one. In B. subtilis, FliY is the functional equivalent of FliN, and both proteins share the dimerization domain. However, FliY contains a CheC-like domain (25) and an N-terminal extension to which FlhG binds (Fig. 6B). Based on these observations, we speculate that the diversity in flagellar C-ring proteins represents a determinant of flagellation pattern control. In C. jejuni, FliN differs from its counterparts in a yet undefined domain that resides N-terminally to its dimerization domain (Fig. 6A). C. jejuni lacks the MinCD system, and, in addition to their canonical role, FliM, FliN, and FlhG are important for inhibiting futile cell division at the cell poles (7). These observations suggest that the diversity of the C-ring components not only is important for flagellation pattern control but also may link other cellular processes to flagella assembly.
Fig. 6.
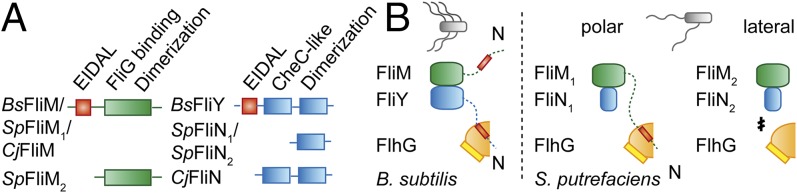
FlhG and the diversity of flagellar C-ring proteins. (A) Diversity in the architecture of the C-ring proteins FliM (Left, green) and FliY/FliN (Right, blue) from B. subtilis, S. putrefaciens, and C. jejuni. The conserved EIDAL motif is shown in red. (B) Interaction of FlhG with flagellar C-ring complexes in B. subtilis (Left) and S. putrefaciens (Right). The EIDAL motifs are shown in red. “N” indicates N termini.
Materials and Methods
Experimental procedures are described in detail in SI Materials and Methods and a strain list is given in Table S2.
Protein Purification.
Proteins from B. subtilis, G. thermodenitrificans, and S. putrefaciens were obtained as described in SI Materials and Methods. Briefly, all proteins were produced in E. coli BL21 (DE3) and were purified by Ni-ion affinity and SEC. The SEC buffer consisted of 20 mM Hepes-Na (pH 7.5), 200 mM NaCl, 20 mM KCl, and 20 mM MgCl2. FliM and FliY were coproduced and purified via the same protocol with a (His)6 tag at the C terminus of FliM.
Crystallization and Structure Determination.
Crystallization was performed by the sitting-drop method at 20 °C as further detailed in SI Materials and Methods. Data were collected at the European Synchrotron Radiation Facility and were processed with iMosflm (41) and SCALA (42). Structures were determined by molecular replacement with PHASER (43), built in COOT (44), and refined with PHENIX (45). Search models were EcMinD (PDB ID code: 3QL9) and GtFlhG (PDB ID code: 4RZ2).
HDX.
HDX is described in detail in SI Materials and Methods. Briefly, purified proteins and their complexes were incubated in deuterated buffer at 37 °C, and the 1H/2H exchange reaction was quenched after 30 s by ice-cold quenching buffer (pH 2.2). Peptic peptides were generated by an online pepsin column and separated by reversed-phase HPLC. Data were analyzed using the HDX workbench (46).
Flotation Assays.
Floatation assays are described in detail in SI Materials and Methods. Briefly, LUVs (PE:PG ratio 70:30) were prepared by extrusion (100-nm pores). FlhG and LUVs were incubated for 10 min at 20 °C and were subjected to iodixanol gradient ultracentrifugation. Proteins were precipitated with trichloroacetic acid (TCA) and analyzed by Coomassie-stained SDS/PAGE.
GST-Binding Assays.
GST assays are described in detail in SI Materials and Methods. All assays were performed in PBS at 4 °C. GST-protein (1 nmol) was immobilized to glutathione-Sepharose beads (GE Healthcare). Next, 2 nmol of a potential interaction partner was incubated for 10 min at 4 °C. Beads were washed with PBS, and GST-proteins or their complexes were eluted with 20 mM glutathione in 50 mM Tris⋅HCl (pH 7.5) and were analyzed by Coomassie-stained SDS/PAGE.
Hydrolysis Assays.
ATPase activity was monitored by HPLC referenced to ADP and ATP standards (see also SI Materials and Methods). Nucleotides were separated on a C18 column (isocratic flow; 0.8 mL/min) with a phosphate buffer containing 10 mM tetrapentylammonium bromide (TPAB) and 15% (vol/vol) acetonitrile. Nucleotides were quantified (by peak area) using ChemStation (B.04.03).
Motility Assays.
Spreading of S. putrefaciens CN-32 or its mutants was monitored by light microscopy or on soft-agar plates, using protocols that were established earlier (28) and are described in detail in SI Materials and Methods.
Fluorescence Microscopy.
Fluorescence microscopy was performed with B. subtilis immobilized on agarose pads at a laser-scanning microscope using a 100× objective. For colocalization, YFP and CFP were recorded simultaneously to allow highest spatial accuracy over time. Microscopy on S. putrefaciens CN-32 was executed as described (28) with a DMI 6000B microscope (Leica) equipped with a 100× objective. Images were collected and processed with the VisiView Premier software (Visitron Systems) and ImageJ 1.47v software (47).
Transmission Electron Microscopy.
Cells were applied to carbon-coated copper grids and negatively stained with 2% (wt/vol) uranyl acetate (27). Electron microscopy was performed on a JEOL JEM-2100 at 120 kV. Further details are given in SI Materials and Methods.
Atomic coordinates and structure factors have been deposited with the Protein Data Bank (PDB) under ID codes 4RZ2 and 4RZ3.
Supplementary Material
Acknowledgments
We thank Irmi Sinning for continuous support; Peter Graumann and Uwe-G. Maier for support with fluorescence and electron microscopy, respectively; Daniel B. Kearns for providing B. subtilis strains; Nico Kümmerer for his contribution in the beginning of the project; and Ulrike Ruppert for technical assistance. We are grateful to Matthew McIntosh for commenting on the manuscript. We acknowledge the assistance of the European Synchrotron Radiation Facility (Grenoble). This project was supported by the LOEWE Program of the State of Hesse (G.B.), the Peter and Traudl Engelhorn Foundation (G.B.), Fonds der Chemischen Industrie (G.B. and J.S.S.), and the Deutsche Forschungsgemeinschaft through TH831/5-1 within the framework SPP1617 (to K.M.T.) and INST 160/621-1 FUGG (to G.B. and U.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org/ (PDB ID codes 4RZ2 and 4RZ3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419388112/-/DCSupplemental.
References
- 1.Chevance FF, Hughes KT. Coordinating assembly of a bacterial macromolecular machine. Nat Rev Microbiol. 2008;6(6):455–465. doi: 10.1038/nrmicro1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altegoer F, Schuhmacher J, Pausch P, Bange G. From molecular evolution to biobricks and synthetic modules: A lesson by the bacterial flagellum. Biotechnol Genet Eng Rev. 2014;30:49–64. doi: 10.1080/02648725.2014.921500. [DOI] [PubMed] [Google Scholar]
- 3.Kusumoto A, et al. Regulation of polar flagellar number by the flhF and flhG genes in Vibrio alginolyticus. J Biochem. 2006;139(1):113–121. doi: 10.1093/jb/mvj010. [DOI] [PubMed] [Google Scholar]
- 4.Campos-García J, Nájera R, Camarena L, Soberón-Chávez G. The pseudomonas aeruginosa motR gene involved in regulation of bacterial motility. FEMS Microbiol Lett. 2000;184(1):57–62. doi: 10.1111/j.1574-6968.2000.tb08990.x. [DOI] [PubMed] [Google Scholar]
- 5.Dasgupta N, Arora SK, Ramphal R. fleN, a gene that regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2000;182(2):357–364. doi: 10.1128/jb.182.2.357-364.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Amsterdam K, van der Ende A. Helicobacter pylori HP1034 (ylxH) is required for motility. Helicobacter. 2004;9(5):387–395. doi: 10.1111/j.1083-4389.2004.00268.x. [DOI] [PubMed] [Google Scholar]
- 7.Balaban M, Hendrixson DR. Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni. PLoS Pathog. 2011;7(12):e1002420. doi: 10.1371/journal.ppat.1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guttenplan SB, Shaw S, Kearns DB. The cell biology of peritrichous flagella in Bacillus subtilis. Mol Microbiol. 2013;87(1):211–229. doi: 10.1111/mmi.12103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukherjee S, Kearns DB. The structure and regulation of flagella in Bacillus subtilis. Annu Rev Genet. 2014;48:319–340. doi: 10.1146/annurev-genet-120213-092406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bange G, Petzold G, Wild K, Parlitz RO, Sinning I. The crystal structure of the third signal-recognition particle GTPase FlhF reveals a homodimer with bound GTP. Proc Natl Acad Sci USA. 2007;104(34):13621–13625. doi: 10.1073/pnas.0702570104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bange G, et al. Structural basis for the molecular evolution of SRP-GTPase activation by protein. Nat Struct Mol Biol. 2011;18(12):1376–1380. doi: 10.1038/nsmb.2141. [DOI] [PubMed] [Google Scholar]
- 12.Kusumoto A, et al. Collaboration of FlhF and FlhG to regulate polar-flagella number and localization in Vibrio alginolyticus. Microbiology. 2008;154(Pt 5):1390–1399. doi: 10.1099/mic.0.2007/012641-0. [DOI] [PubMed] [Google Scholar]
- 13.Schniederberend M, Abdurachim K, Murray TS, Kazmierczak BI. The GTPase activity of FlhF is dispensable for flagellar localization, but not motility, in Pseudomonas aeruginosa. J Bacteriol. 2013;195(5):1051–1060. doi: 10.1128/JB.02013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parrish JR, et al. A proteome-wide protein interaction map for Campylobacter jejuni. Genome Biol. 2007;8(7):R130. doi: 10.1186/gb-2007-8-7-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Green JC, et al. Recruitment of the earliest component of the bacterial flagellum to the old cell division pole by a membrane-associated signal recognition particle family GTP-binding protein. J Mol Biol. 2009;391(4):679–690. doi: 10.1016/j.jmb.2009.05.075. [DOI] [PubMed] [Google Scholar]
- 16.Leipe DD, Wolf YI, Koonin EV, Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J Mol Biol. 2002;317(1):41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 17.Lutkenhaus J. The ParA/MinD family puts things in their place. Trends Microbiol. 2012;20(9):411–418. doi: 10.1016/j.tim.2012.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lutkenhaus J, Pichoff S, Du S. Bacterial cytokinesis: From Z ring to divisome. Cytoskeleton (Hoboken) 2012;69(10):778–790. doi: 10.1002/cm.21054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu W, Park KT, Holyoak T, Lutkenhaus J. Determination of the structure of the MinD-ATP complex reveals the orientation of MinD on the membrane and the relative location of the binding sites for MinE and MinC. Mol Microbiol. 2011;79(6):1515–1528. doi: 10.1111/j.1365-2958.2010.07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szeto TH, Rowland SL, Habrukowich CL, King GF. The MinD membrane targeting sequence is a transplantable lipid-binding helix. J Biol Chem. 2003;278(41):40050–40056. doi: 10.1074/jbc.M306876200. [DOI] [PubMed] [Google Scholar]
- 21.Zhou H, Lutkenhaus J. Membrane binding by MinD involves insertion of hydrophobic residues within the C-terminal amphipathic helix into the bilayer. J Bacteriol. 2003;185(15):4326–4335. doi: 10.1128/JB.185.15.4326-4335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dajkovic A, Lan G, Sun SX, Wirtz D, Lutkenhaus J. MinC spatially controls bacterial cytokinesis by antagonizing the scaffolding function of FtsZ. Curr Biol. 2008;18(4):235–244. doi: 10.1016/j.cub.2008.01.042. [DOI] [PubMed] [Google Scholar]
- 23.Leonard TA, Butler PJ, Löwe J. Bacterial chromosome segregation: Structure and DNA binding of the Soj dimer—a conserved biological switch. EMBO J. 2005;24(2):270–282. doi: 10.1038/sj.emboj.7600530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stjepanovic G, et al. Lipids trigger a conformational switch that regulates signal recognition particle (SRP)-mediated protein targeting. J Biol Chem. 2011;286(26):23489–23497. doi: 10.1074/jbc.M110.212340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szurmant H, Bunn MW, Cannistraro VJ, Ordal GW. Bacillus subtilis hydrolyzes CheY-P at the location of its action, the flagellar switch. J Biol Chem. 2003;278(49):48611–48616. doi: 10.1074/jbc.M306180200. [DOI] [PubMed] [Google Scholar]
- 26.Kazmierczak BI, Hendrixson DR. Spatial and numerical regulation of flagellar biosynthesis in polarly flagellated bacteria. Mol Microbiol. 2013;88(4):655–663. doi: 10.1111/mmi.12221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bubendorfer S, et al. Specificity of motor components in the dual flagellar system of Shewanella putrefaciens CN-32. Mol Microbiol. 2012;83(2):335–350. doi: 10.1111/j.1365-2958.2011.07934.x. [DOI] [PubMed] [Google Scholar]
- 28.Bubendorfer S, Koltai M, Rossmann F, Sourjik V, Thormann KM. Secondary bacterial flagellar system improves bacterial spreading by increasing the directional persistence of swimming. Proc Natl Acad Sci USA. 2014;111(31):11485–11490. doi: 10.1073/pnas.1405820111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray TS, Kazmierczak BI. FlhF is required for swimming and swarming in Pseudomonas aeruginosa. J Bacteriol. 2006;188(19):6995–7004. doi: 10.1128/JB.00790-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science. 2008;320(5883):1636–1638. doi: 10.1126/science.1157877. [DOI] [PubMed] [Google Scholar]
- 31.Courtney CR, Cozy LM, Kearns DB. Molecular characterization of the flagellar hook in Bacillus subtilis. J Bacteriol. 2012;194(17):4619–4629. doi: 10.1128/JB.00444-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lackner LL, Raskin DM, de Boer PA. ATP-dependent interactions between Escherichia coli Min proteins and the phospholipid membrane in vitro. J Bacteriol. 2003;185(3):735–749. doi: 10.1128/JB.185.3.735-749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park KT, et al. The Min oscillator uses MinD-dependent conformational changes in MinE to spatially regulate cytokinesis. Cell. 2011;146(3):396–407. doi: 10.1016/j.cell.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park KT, Wu W, Lovell S, Lutkenhaus J. Mechanism of the asymmetric activation of the MinD ATPase by MinE. Mol Microbiol. 2012;85(2):271–281. doi: 10.1111/j.1365-2958.2012.08110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dasgupta N, Ramphal R. Interaction of the antiactivator FleN with the transcriptional activator FleQ regulates flagellar number in Pseudomonas aeruginosa. J Bacteriol. 2001;183(22):6636–6644. doi: 10.1128/JB.183.22.6636-6644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Correa NE, Peng F, Klose KE. Roles of the regulatory proteins FlhF and FlhG in the Vibrio cholerae flagellar transcription hierarchy. J Bacteriol. 2005;187(18):6324–6332. doi: 10.1128/JB.187.18.6324-6332.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jain R, Kazmierczak BI. A conservative amino acid mutation in the master regulator FleQ renders Pseudomonas aeruginosa aflagellate. PLoS ONE. 2014;9(5):e97439. doi: 10.1371/journal.pone.0097439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bren A, Eisenbach M. The N terminus of the flagellar switch protein, FliM, is the binding domain for the chemotactic response regulator, CheY. J Mol Biol. 1998;278(3):507–514. doi: 10.1006/jmbi.1998.1730. [DOI] [PubMed] [Google Scholar]
- 39.Welch M, Oosawa K, Aizawa S, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90(19):8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sourjik V, Armitage JP. Spatial organization in bacterial chemotaxis. EMBO J. 2010;29(16):2724–2733. doi: 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: A new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67(Pt 4):235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40(Pt 4):658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 2):213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pascal BD, et al. HDX workbench: Software for the analysis of H/D exchange MS data. J Am Soc Mass Spectrom. 2012;23(9):1512–1521. doi: 10.1007/s13361-012-0419-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012;9(7):671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.